Abstract
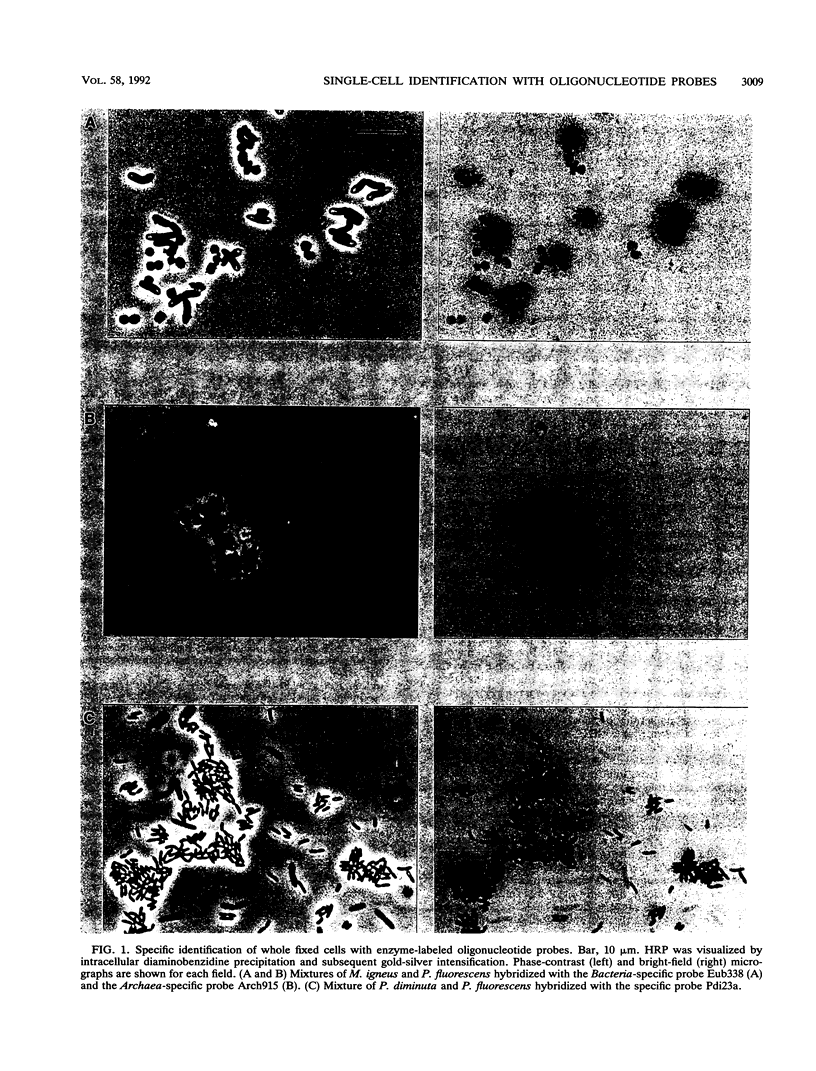
A method to microscopically detect and identify individual cells of members of the domains Bacteria and Archaea is presented. rRNA-targeted oligonucleotides were 5' end labeled with the enzyme horseradish peroxidase and used for whole-cell hybridization. Specifically bound probe was visualized by the enzymatic formation of an intracellular precipitate from the substrate diaminobenzidine. Permeation of the enzyme-labeled probe into whole fixed cells of gram-negative bacteria required their pretreatment with lysozyme-EDTA, whereas permeability of some archaebacterial cells was improved by addition of detergent to the hybridization buffer. Hitherto we had not achieved penetration of enzyme-labeled probe into gram-positive bacteria and yeast cells. This method should be a valuable tool for identification of suitable prokaryotic cells in environments with elevated background fluorescence or in situations in which an epifluorescence microscope is not available.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Bronstein I., McGrath P. Chemiluminescence lights up. Nature. 1989 Apr 13;338(6216):599–600. doi: 10.1038/338599a0. [DOI] [PubMed] [Google Scholar]
- DeLong E. F., Wickham G. S., Pace N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989 Mar 10;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- Doddema H. J., Vogels G. D. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1978 Nov;36(5):752–754. doi: 10.1128/aem.36.5.752-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallyas F., Görcs T., Merchenthaler I. High-grade intensification of the end-product of the diaminobenzidine reaction for peroxidase histochemistry. J Histochem Cytochem. 1982 Feb;30(2):183–184. doi: 10.1177/30.2.7061820. [DOI] [PubMed] [Google Scholar]
- Graham R. C., Jr, Karnovsky M. J. The early stages of absorption of injected horseradish peroxidase in the proximal tubules of mouse kidney: ultrastructural cytochemistry by a new technique. J Histochem Cytochem. 1966 Apr;14(4):291–302. doi: 10.1177/14.4.291. [DOI] [PubMed] [Google Scholar]
- Jablonski E., Moomaw E. W., Tullis R. H., Ruth J. L. Preparation of oligodeoxynucleotide-alkaline phosphatase conjugates and their use as hybridization probes. Nucleic Acids Res. 1986 Aug 11;14(15):6115–6128. doi: 10.1093/nar/14.15.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Medon P. P., Skingle D. C., Lanser J. A., Symons R. H. Enzyme-linked synthetic oligonucleotide probes: non-radioactive detection of enterotoxigenic Escherichia coli in faecal specimens. Nucleic Acids Res. 1987 Jul 10;15(13):5275–5287. doi: 10.1093/nar/15.13.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHAECHTER M., MAALOE O., KJELDGAARD N. O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958 Dec;19(3):592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Scopsi L., Larsson L. I. Increased sensitivity in peroxidase immunocytochemistry. A comparative study of a number of peroxidase visualization methods employing a model system. Histochemistry. 1986;84(3):221–230. doi: 10.1007/BF00495786. [DOI] [PubMed] [Google Scholar]
- Tsien H. C., Bratina B. J., Tsuji K., Hanson R. S. Use of oligodeoxynucleotide signature probes for identification of physiological groups of methylotrophic bacteria. Appl Environ Microbiol. 1990 Sep;56(9):2858–2865. doi: 10.1128/aem.56.9.2858-2865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdea M. S., Warner B. D., Running J. A., Stempien M., Clyne J., Horn T. A comparison of non-radioisotopic hybridization assay methods using fluorescent, chemiluminescent and enzyme labeled synthetic oligodeoxyribonucleotide probes. Nucleic Acids Res. 1988 Jun 10;16(11):4937–4956. doi: 10.1093/nar/16.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarda B., Amann R., Wallner G., Schleifer K. H. Identification of single bacterial cells using digoxigenin-labelled, rRNA-targeted oligonucleotides. J Gen Microbiol. 1991 Dec;137(12):2823–2830. doi: 10.1099/00221287-137-12-2823. [DOI] [PubMed] [Google Scholar]