Abstract
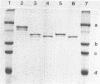
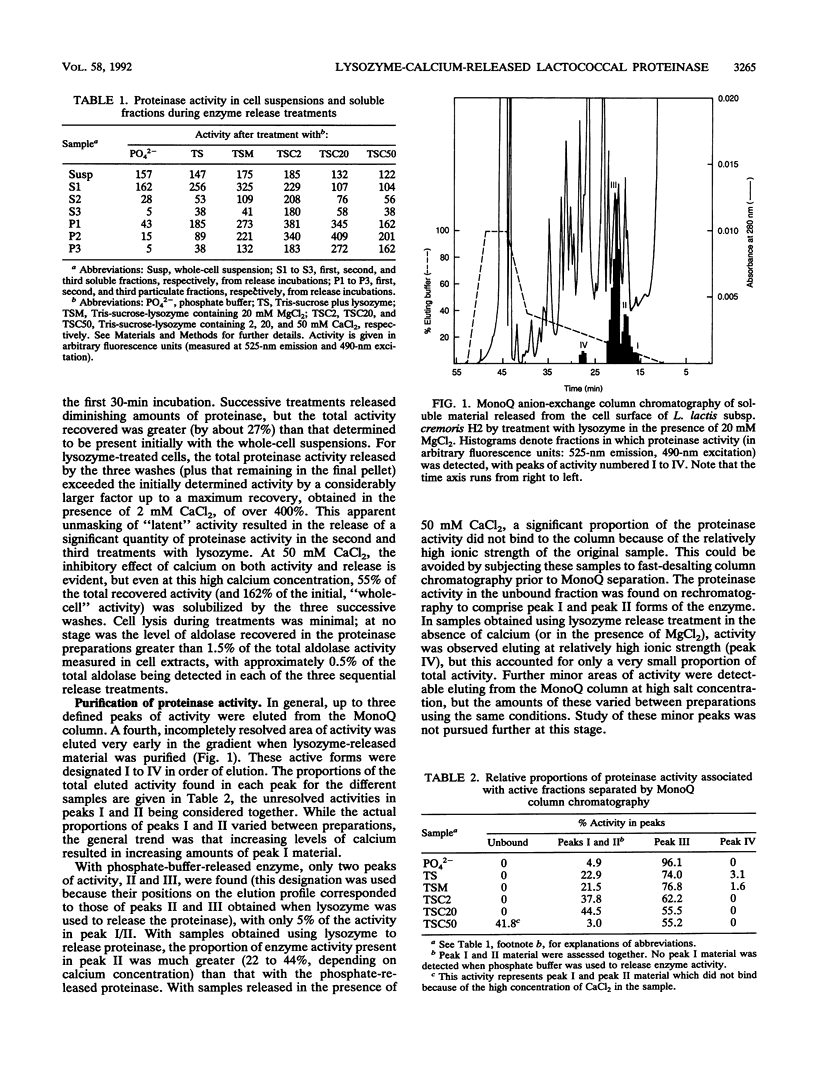
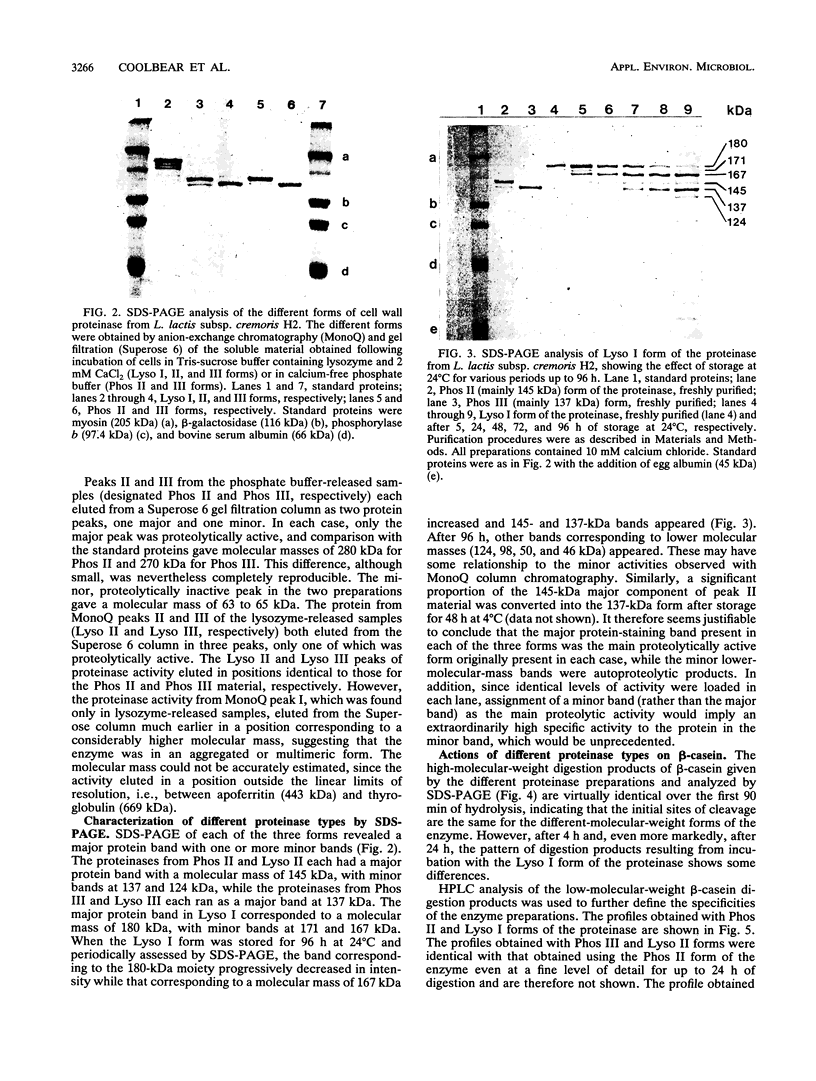
The cell wall-associated proteinase from Lactococcus lactis subsp. cremoris H2 (isolate number 4409) was released from the cells by treatment with lysozyme, even in the presence of 50 mM calcium chloride. Cell lysis during lysozyme treatment was minimal. The proteinase activity released by lysozyme treatment fractionated on ion-exchange chromatography as three main forms, the molecular masses of which were determined by gel exclusion chromatography and polyacrylamide gel electrophoresis. Two of the enzyme forms released, 137 and 145 kDa, were the same as those released by incubation of cells in calcium-free phosphate buffer. In the presence of calcium, lysozyme treatment also resulted in the release of a 180-kDa enzyme molecule. The total proteinase activity released by lysozyme treatment (in the presence or absence of calcium) was not only greater than that released by phosphate buffer but was also greater than that initially detectable on the surface of whole cells, suggesting an unmasking of enzyme on the cell surface. The presence of calcium during release treatment resulted in increased stability of the crude enzyme preparations. For the proteinase preparation released by using lysozyme with 50 mM CaCl2, the half-life of proteinase activity at 37°C was 39 h, compared with 0.22 h for the calcium-free phosphate buffer-released preparation. In all cases, maximum stability was observed at pH 5.5. Comparison of β-casein hydrolysis by the three forms of the enzyme showed that the products of short-term (5- to 30-min) digestions were very similar, although subtle differences were detected with the 180-kDa form.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Crow V. L., Thomas T. D. D-tagatose 1,6-diphosphate aldolase from lactic streptococci: purification, properties, and use in measuring intracellular tagatose 1,6-diphosphate. J Bacteriol. 1982 Aug;151(2):600–608. doi: 10.1128/jb.151.2.600-608.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz J., Exterkate F., Konings W. N. The Proteolytic Systems of Streptococcus cremoris: an Immunological Analysis. Appl Environ Microbiol. 1984 Dec;48(6):1105–1110. doi: 10.1128/aem.48.6.1105-1110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwaki M., Ikemura H., Shimizu-Kadota M., Hirashima A. Molecular characterization of a cell wall-associated proteinase gene from Streptococcus lactis NCDO763. Mol Microbiol. 1989 Mar;3(3):359–369. doi: 10.1111/j.1365-2958.1989.tb00181.x. [DOI] [PubMed] [Google Scholar]
- Kok J., Hill D., Haandrikman A. J., de Reuver M. J., Laan H., Venema G. Deletion analysis of the proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):239–244. doi: 10.1128/aem.54.1.239-244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan H., Konings W. N. Autoproteolysis of the Extracellular Serine Proteinase of Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1991 Sep;57(9):2586–2590. doi: 10.1128/aem.57.9.2586-2590.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan H., Konings W. N. Mechanism of Proteinase Release from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1989 Dec;55(12):3101–3106. doi: 10.1128/aem.55.12.3101-3106.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan H., Smid E. J., de Leij L., Schwander E., Konings W. N. Monoclonal Antibodies to the Cell-Wall-Associated Proteinase of Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1988 Sep;54(9):2250–2256. doi: 10.1128/aem.54.9.2250-2256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Monnet V., Le Bars D., Gripon J. C. Purification and characterization of a cell wall proteinase from Streptococcus lactis NCDO 763. J Dairy Res. 1987 May;54(2):247–255. doi: 10.1017/s0022029900025383. [DOI] [PubMed] [Google Scholar]
- Nissen-Meyer J., Sletten K. Purification and characterization of the free form of the lactococcal extracellular proteinase and its autoproteolytic cleavage products. J Gen Microbiol. 1991 Jul;137(7):1611–1618. doi: 10.1099/00221287-137-7-1611. [DOI] [PubMed] [Google Scholar]
- Reid J. R., Moore C. H., Midwinter G. G., Pritchard G. G. Action of a cell wall proteinase from Lactococcus lactis subsp. cremoris SK11 on bovine alpha s1-casein. Appl Microbiol Biotechnol. 1991 May;35(2):222–227. doi: 10.1007/BF00184690. [DOI] [PubMed] [Google Scholar]
- Reid J. R., Ng K. H., Moore C. H., Coolbear T., Pritchard G. G. Comparison of bovine beta-casein hydrolysis by PI and PIII-type proteinases from Lactococcus lactis subsp. cremoris [corrected]. Appl Microbiol Biotechnol. 1991 Dec;36(3):344–351. doi: 10.1007/BF00208154. [DOI] [PubMed] [Google Scholar]
- Siezen R. J., de Vos W. M., Leunissen J. A., Dijkstra B. W. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991 Oct;4(7):719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- Twining S. S. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal Biochem. 1984 Nov 15;143(1):30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- Visser S., Exterkate F. A., Slangen C. J., de Veer G. J. Comparative Study of Action of Cell Wall Proteinases from Various Strains of Streptococcus cremoris on Bovine alpha(s1)-, beta-, and kappa-Casein. Appl Environ Microbiol. 1986 Nov;52(5):1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S., Robben A. J., Slangen C. J. Specificity of a cell-envelope-located proteinase (PIII-type) from Lactococcus lactis subsp. cremoris AM1 in its action on bovine beta-casein. Appl Microbiol Biotechnol. 1991 Jul;35(4):477–483. doi: 10.1007/BF00169753. [DOI] [PubMed] [Google Scholar]
- Vos P., Simons G., Siezen R. J., de Vos W. M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989 Aug 15;264(23):13579–13585. [PubMed] [Google Scholar]