Abstract
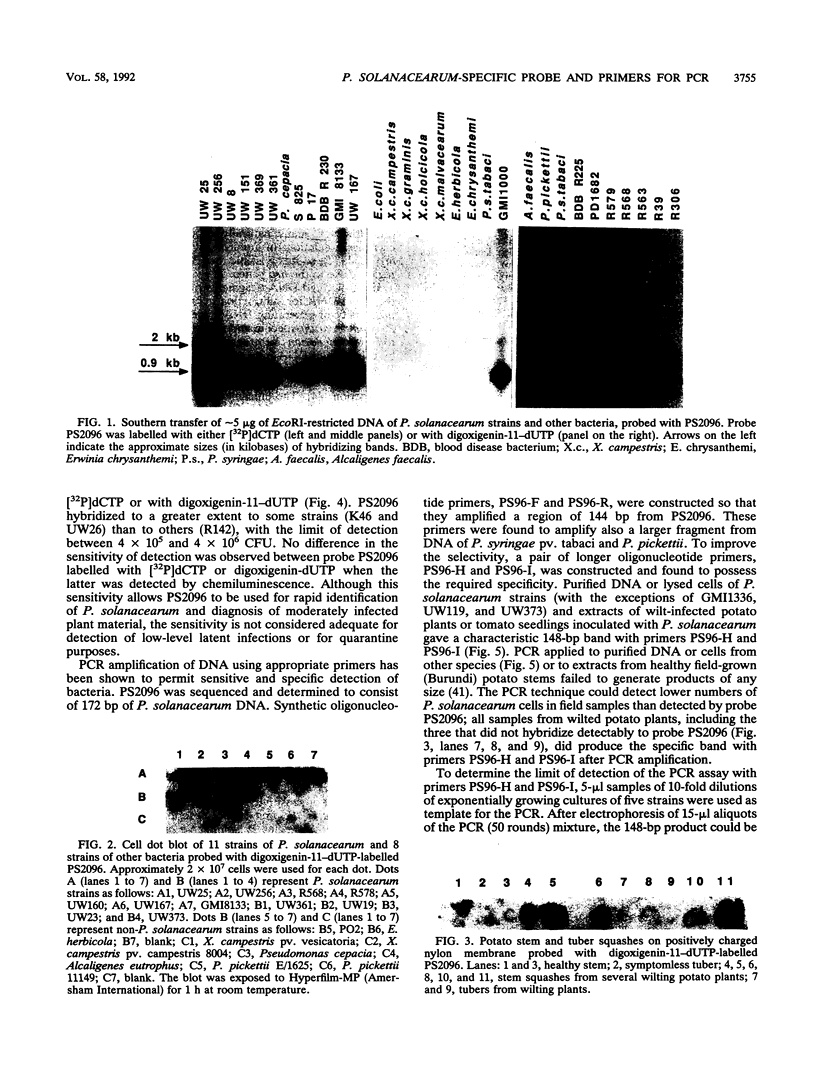
A subtraction hybridization technique was employed to make a library enriched for Pseudomonas solanacearum-specific sequences. One cloned fragment, PS2096, hybridized under stringent conditions to DNA of 82 P. solanacearum strains representing all subgroups of the species. Other plant-associated bacteria, including closely related species such as Pseudomonas capacia, Pseudomonas picketti, or Pseudomonas syzygii, did not hybridize to PS2096. A minimum number of between 4 x 10(5) and 4 x 10(6) P. solanacearum cells could routinely be detected with PS2096 labelled either with [32P]dCTP or with digoxigenin-11-dUTP. To improve the sensitivity of detection, PS2096 was sequenced to allow the construction of specific oligonucleotide primers to be used for polymerase chain reaction (PCR) amplification. After 50 cycles of amplification, 5 to 116 cells, depending on the strain, could reproducibly be detected by visualization of a 148-bp PCR product on an agarose gel. A preliminary field trial in Burundi with the probe and PCR primers has confirmed that they are sensitive tools for specifically detecting low-level infections of P. solanacearum in potato tubers.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avery R. J., Norton J. D., Jones J. S., Burke D. C., Morris A. G. Interferon inhibits transformation by murine sarcoma viruses before integration of provirus. Nature. 1980 Nov 6;288(5786):93–95. doi: 10.1038/288093a0. [DOI] [PubMed] [Google Scholar]
- Bochner B. R., Savageau M. A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977 Feb;33(2):434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher C. A., Van Gijsegem F., Barberis P. A., Arlat M., Zischek C. Pseudomonas solanacearum genes controlling both pathogenicity on tomato and hypersensitivity on tobacco are clustered. J Bacteriol. 1987 Dec;169(12):5626–5632. doi: 10.1128/jb.169.12.5626-5632.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D., Sequeira L. The use of subtractive hybridization to obtain a DNA probe specific for Pseudomonas solanacearum race 3. Mol Gen Genet. 1991 Jul;227(3):401–410. doi: 10.1007/BF00273930. [DOI] [PubMed] [Google Scholar]
- Deneer H. G., Boychuk I. Species-specific detection of Listeria monocytogenes by DNA amplification. Appl Environ Microbiol. 1991 Feb;57(2):606–609. doi: 10.1128/aem.57.2.606-609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitts R., Diamond M., Hamilton C., Neri M. DNA-DNA hybridization assay for detection of Salmonella spp. in foods. Appl Environ Microbiol. 1983 Nov;46(5):1146–1151. doi: 10.1128/aem.46.5.1146-1151.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont P. A., Grimont F., Desplaces N., Tchen P. DNA probe specific for Legionella pneumophila. J Clin Microbiol. 1985 Mar;21(3):431–437. doi: 10.1128/jcm.21.3.431-437.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. E., Payne W. L., Aulisio C. C. Detection and enumeration of virulent Yersinia enterocolitica in food by DNA colony hybridization. Appl Environ Microbiol. 1983 Sep;46(3):636–641. doi: 10.1128/aem.46.3.636-641.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus D., Ausubel F. M. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1889–1893. doi: 10.1073/pnas.87.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson B. H. Different temperature optima for methane formation when enrichments from Acid peat are supplemented with acetate or hydrogen. Appl Environ Microbiol. 1984 Aug;48(2):389–394. doi: 10.1128/aem.48.2.389-394.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood P. K., Morris J. G., Jr, Small P. L., Sethabutr O., Toledo M. R., Trabulsi L., Kaper J. B. Comparison of DNA probes and the Sereny test for identification of invasive Shigella and Escherichia coli strains. J Clin Microbiol. 1986 Sep;24(3):498–500. doi: 10.1128/jcm.24.3.498-500.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]