Abstract
In invertebrates and vertebrates, carotenoids are ubiquitous colorants, antioxidants, and provitamin A compounds that must be absorbed from dietary sources and transported to target tissues where they are taken up and stabilized to perform their physiological functions. These processes occur in a specific and regulated manner mediated by high-affinity carotenoid-binding proteins. In this mini-review, we examine the published literature on carotenoid-binding proteins in vertebrate and invertebrate systems, and we report our initial purification and characterization of a novel lutein-binding protein isolated from liver of Japanese quail (Coturnix japonica).
Keywords: Carotenoid-binding Proteins, Crustacyanin, GSTP1, StAR protein, Lutein-binding protein, Japanese quail
INTRODUCTION
Carotenoids are omnipresent and exceedingly colored pigments synthesized exclusively by flora and microbes where they perform critical functions in photosynthesis and photoprotection [1,2,3]. They are lipophilic biomolecules and are classified as carotenes if their sole constituents are hydrocarbons or xanthophylls if they also contain one or more oxygen atoms. Over 600 carotenoids have been isolated from natural sources [1]. Vertebrates and invertebrates cannot synthesize carotenoids, but they can ingest them in their diet and subsequently utilize them for important physiological functions [4,5]. The earliest role established for carotenes in animals was as a vitamin A precursor, typically by way of cleavage of the polyene carotenoid backbone to form one or two functional retinoid molecules. Many different carotenoids can be metabolized to products with retinoid activity [6,7]; however, because of the presence of oxygenated groups in terminal ionone rings, most xanthophylls do not fit the structural requirements for provitamin A activity so they serve other non-vitamin A functions [3,6]. Along with carotenes, oxygenated carotenoids are efficient antioxidants that can quench singlet oxygen, reactive oxygen species, and various other free radicals that are by-products of metabolic process in cells or from environmental pollutants [8,9,10]. They also can function as photoprotectants against light-induced damage in heavily exposed tissues such as skin and the eye [11,12].
Humans typically consume a wide variety of carotenoids in the diet, yet β-carotene along with lycopene, lutein, zeaxanthin, β-cryptoxanthin and α-carotene account for 90% of circulating carotenoids [13]. Other carotenoids such as violaxanthin and neoxanthin are common in the human diet but do not seem to be absorbed at all, implying that the first level of uptake specificity is occurring in the gut. Deming et al. have explored the mechanisms of carotenoid absorption and metabolism in the mammalian digestive tract [14]. Carotenes and unesterified xanthophylls are absorbed by mucosal cells and subsequently appear unchanged in the circulation and peripheral tissue, while esterified xanthophylls are cleaved prior to uptake [13,15]. Once in the circulation, the absorbed carotenoids are bound to lipoproteins and then targeted to a variety of tissues such as liver, macula, lung, adipose, brain, prostate, and skin [13, 16]. Some tissues such as skin are relatively nonselective--their carotenoid levels and compositions largely reflect that of the serum [16] while other tissues such as the macula of the human eye are extraordinarily selective, concentrating very high levels of just lutein and zeaxanthin and their metabolites [17–19]. The mechanisms for high-affinity selective uptake of carotenoids are not yet fully understood, but carotenoid-binding proteins are likely to play an important role in dictating tissue specific accumulation [20–21]. These binding proteins may act as cell surface receptors, as transmembrane transport proteins, as metabolic enzymes, as intracellular mediators of the biological actions of the ligand, or as sites for the deposition and stabilization of the ligand. Carotenoid-binding proteins have been extensively described in plants [22] and micro-organisms [23] which synthesize them and utilize them in photosynthesis and photoprotection [24–27]. A few carotenoid-binding proteins have been well studied in invertebrates [28–30], but relatively little information has been available about specific carotenoid-binding proteins in any vertebrate systems. In this mini-review, we assemble published literature of carotenoid binding proteins in vertebrate and invertebrate systems, and we report our initial purification and characterization of a novel lutein-binding protein from liver of Japanese quail (Coturnix japonica).
INVERTEBRATE CAROTENOID-BINDING PROTEINS
Among insects, the silkworm, Bombyx mori, has had its carotenoid-binding proteins most extensively studied because carotenoids impart various colors to their cocoons which in wild-type B. mori are yellow in color due to the presence of lutein. The mechanism by which carotenoids are transported from the lumen of the midgut to the hemolymph lipoprotein, lipophorin, and from lipophorin into the silk gland, where the cocoon is produced, is the primary focus. Jouni and Wells in 1996 initially reported a specific lutein-binding protein (LBP) from the midgut of B. mori [29]. The absorption spectrum of this purified lutein-binding protein was characterized by three absorbance maxima in the visible region at 432, 460, and 492 nm which was a significant red shift of 22–38 nm compared with the spectrum of lutein in hexane or acetone [29]. Although purified to apparent homogeneity as a 35 kDa protein, its sequence was not reported. Once absorbed from the midgut, lutein in the silkworm then becomes associated with high density lipophorin (HDLp) in the hemolymph, a lipoprotein that appears to mediate carotenoid transport in a manner similar to mammalian HDL. Directed tissue transfer is facilitated by the lipophorin receptor and by a lipid transfer particle (LTP) protein which appears necessary for exchange of lutein between various lipophorin molecules [31–32].
In 2002, Tabunoki and colleagues studied a series of mutant silkworms with colorless cocoons, and reported the isolation and cDNA sequence of a 33 kDa lutein-specific carotenoid binding protein (CBP) from the midgut epithelium and silk gland [28]. This protein was a new member of the steroidogenic acute regulatory (StAR) family usually involved in cellular cholesterol transport. It is able to specifically and stoichiometrically bind lutein with a ratio of 1 mol of lutein per 1 mol of CBP. Sequence identity of B. mori CBP and mouse MLN64 and human StAR protein was 29% and 25%, respectively, and high levels of StAR have been reported in carotenoid-rich tissues such as human corpus luteum [33–34]. The absorption spectrum of B. mori CBP was characterized by three absorbance maxima in the visible region at 436, 461, and 493 nm, a significant red shift of 22 nm compared with lutein’s spectrum in hexane Its spectrum was very close to the reported absorption spectra of LBP isolated from the midgut of B. mori by Jouni and Wells [29,35], suggesting that silkworm LBP and CBP might be the same protein, although their reported stoichiometries of binding differ with a lutein to protein ratio of 3:1 for LBP and 1:1 for CBP. Functional characterization studies by microscopic immunocytochemistry clearly indicated that this protein might be involved in aiding absorption and uptake of carotenoids from lipophorin [35] which is ultimately responsible for pigmentation of the yellow cocoons [36].
Another well studied invertebrate carotenoid binding protein is crustacyanin isolated from carapace of the lobster Homarus gammarus. Lobster shells are deep blue in color because the xanthophyll carotenoid astaxanthin binds to a carrier protein called crustacyanin. The binding of astaxanthin to crustacyanin results in a bathochromic shift of 160 nm (λmax 632 nm) as compared to the wavelength maximum of unbound astaxanthin (λmax = 472 nm). Denaturation of crustacyanin by boiling accounts for the characteristic color change of lobsters during cooking. The biophysical basis of the very large protein-induced bathochromic shift of astaxanthin by crustacyanin has been a topic of prolific research for over 60 years [37]. Earlier researchers have attributed this binding behavior to interactions with a particular tryptophan residue on the protein molecule [38]. It was also proposed that exciton coupling due to the proximity of the astaxanthin chromophores in the excited state might be the reason for this strong binding and bathochromic shift. This hypothesis was strongly supported by circular dichroism studies with crustacyanin [39, 40]. Recently, this hypothesis was supported by solid-state NMR spectroscopy and resonance Raman studies [41].
α-Crustacyanin was extracted from lobster carapace and subsequently purified by anion-exchange and gel-filtration chromatography by Zagalsky [42]. Crustacyanin has a molecular weight of about 320 kDa, and it is a multi-macromolecular 16-mer complex of protein subunits with 16 bound astaxanthin molecules. It was proposed that α-crustacyanin consists of two groups of homologous proteins (CRTC, containing apoproteins C1, C2 and A1, and CRTA, containing apoproteins A2 and A3) [43]. Cianci et al. 2002 further reported the structural basis of the bathochromic shift and binding behavior based on subunits A1 and A3 of curstacyanin protein and several different perturbations of the carotenoid molecules [44].
Combinations of pairs of these subunits, one from each group, are called β-crustacyanins; they occur naturally in lobster shell and are formed irreversibly by dissociation of α-crustacyanin. The absorption spectrum of the carotenoid astaxanthin along with the β-crustacyanins displays a bathochromic shift of 100 nm (λmax = 580 nm [40]. It was proposed that extension of conjugation by coplanarization of the β-ionone rings with the polyene chain and polarization resulting from hydrogen bonding at the C(4) and C(4′) keto groups of astaxanthin may be mainly responsible for the bathochromic shift [30, 44].
The functional role for crustacyanin is most likely for protective coloration, but antioxidant effects must also be considered. The biochemical properties of silkworm LBP and lobster crustacyanin are summarized in Table 1 in comparison to the vertebrate carotenoid-binding proteins discussed below.
Table 1.
Invertebrate and vertebrate tissue carotenoid-binding proteins and their biochemical properties.
| Source | Major Carotenoid | Mol wt | Bathochromic Shift | Protein | Key reference |
|---|---|---|---|---|---|
| Silkworm silk gland | Lutein | 35 kDa | 22 nm | LBP/BmStart1 | Tabunoki et al. 2002 [28]. |
| Lobster carapace | Astaxanthin | 66 kDa | 160 nm | Crustacyanin | Zagalsky et al. 1991 [38]. |
| Atlantic salmon | Astaxanthin | 109; 105; 95 kDa | NA | Alpha-actinin | Matthews et al. 2006 [69]. |
| Human macula | Zeaxanthin | 23 kDa | 12–20 nm | GSTP1 | Bhosale et al 2004 [21]. |
| Ferret liver | Beta-carotene | 67 kDa | 32 nm | Not yet known | Rao et al. 1997 [78]. |
| Quail liver | Lutein | ~55 kDa | 80 nm | Not yet known | This report |
NA: Information not avaliable
CAROTENOID METABOLIC ENZYMES
Vertebrates and invertebrates generally exhibit very limited capacity to metabolize carotenoids enzymatically, and relatively few carotenoid-specific metabolic enzymes have been reported [42–43]. Nonspecific esterases likely mediate xanthophyll ester cleavage in the gut, and some cells such as bird photoreceptors and frog retinal pigment epithelium re-esterify xanthophylls delivered to the tissue [17,45] Unlike mammals, birds possess a robust ability to add and remove oxygens on various carotenoids and to oxidize and reduce xanthophylls, presumably by enzymatically mediated pathways [46]. meso-Zeaxanthin is a nondietary carotenoid that is found in large amounts uniquely in the eye of primates and birds. Its dietary precursor is lutein, but no enzyme capable of mediating this reaction has been reported [17,21].
On the other hand, in recent years there has been a wealth of new information on the enzymatic basis for the cleavage of carotenoids to form retinoids and apocarotenoids. It had long been speculated that vitamin A production was meditated by a β-β-carotene 15,15′ cleavage enzyme, forming two molecules of retinal from one molecule of beta-carotene by central cleavage, but others argued that eccentric cleavage was the primary event followed by further metabolic processing to form vitamin A [47] . Although beta-carotene cleavage activity was demonstrated in cell-free extracts derived from mammalian small intestine in the 1960s, purification of the enzyme proved surprisingly challenging [47–50]. It was not until the advent of molecular biological approaches and mass spectral sequencing that the beta-carotene cleavage enzyme was cloned first from Drosophila and chicken and later from humans [54–58]. It is a cytosolic enzyme primarily localized in the duodenal mucosa although it has been found in liver and in the eye. It is a 66 kDa sulfhydryl protein, requires molecular oxygen, and is activated by ferrous ions. It has been demonstrated that this enzyme can act on other carotenoids also; however, provitamin A activity might vary depending on the substrate. Cloning of mammalian proteins similar to the 15, 15′ β-carotene oxygenase has revealed the existence of RPE65 [54,56], a protein critical to the cis-trans isomerization of retinoids in the visual cycle, as well as an enzyme that catalyzes the asymmetric cleavage of beta-carotene at the 9′-10′ position [47–49, 57].
PLASMA CAROTENOID BINDING PROTEINS
Plasma lipoproteins are best known as proteins mediating cholesterol transfer between tissues, but Krinsky et al. were the first to propose a further role of lipoproteins in the circulation, transporting of carotenoids [58]. Khachik et al. have identified and quantified as many as 34 major and minor carotenoids including 8 metabolites in the extracts from human plasma and have proposed possible metabolic pathways leading to carotenoid metabolites [59, 60]. The uneven distribution of carotenoids and xanthophylls among the various plasma lipoproteins, chylomicrons, VLDL, LDL, and HDL was then quantified. Chylomicrons mediate the transfer of absorbed carotenoids from the gut to the liver where they are repackaged onto the serum lipoproteins. The stereochemistry of carotenoids may play an important role in their intestinal absorption and cellular uptake and incorporation into chylomicrons. It was proposed that cis-isomers of carotenoids are more bioavailable than trans-carotenoids because cis-isomers are more soluble in bile acid micelles and may be preferentially incorporated into chylomicrons [61].
Non-polar carotenoids such as β-carotene and lycopene are associated primarily with VLDL and LDL, while more polar xanthophylls such as lutein and zeaxanthin prefer to associate with HDL [62]. Once loaded with carotenoids, the apolipoprotein components of lipoproteins are thought to help direct the carotenoids to their target tissues via their interactions with specific cell surface receptors. For example, the beta-carotenoid-rich corpus luteum has abundant LDL receptors [63], and class B scavenger receptors are thought to mediate xanthophyll delivery to ocular tissues in Drosophila (ninaD) and vertebrates (SR-BI) [64].
The association between lipoprotein and carotenoid is relatively non-specific but mutually beneficial [21, 65]. Lipoproteins play an important role in translocation of carotenoids in the mammalian body, and in return carotenoids effectively inhibit oxidation of lipoproteins promoted by reactive molecules. LDL oxidation is generally due to a free radical driven chain reaction wherein polyunsaturated fatty acids are converted to lipid peroxides which easily decompose to many products, including biologically active aldehydes. Carotenoids effectively quench these free radicals to protect and maintain the balance of the LDL and HDL pools in the serum.
In addition to HDL and LDL, albumin, and β-lactoglobulin can act as carrier proteins for carotenoids, although there is no evidence that they do so with high specificity or affinity. Albumin is thought to be a minor carrier of carotenoids in the serum, but it is the major carrier of related compounds such as retinoic acid. Recent reports indicates that fatty acid-free human serum albumin (HSA) may have specific carotenoid binding sites for carotenoids such as lutein and/or meso-astaxanthin [21,66]. β-Lactoglobulin is considered to be a major carotenoid carrier in milk. Its utility as a retinoid and fatty acid binder has been well established and has been studied using spectroscopic techniques [67].
OCULAR CAROTENOID-BINDING PROTEINS
While over 15 different dietary carotenoids are detectable in human serum, only lutein [(3R, 3′R, 6′R)-β, ε-carotene-3,3′-diol] and zeaxanthin [a mixture of (3R,3′R)-β, β-carotene-3,3′-diol and (3R,3′S; meso)-β, β-carotene-3,3′-diol] and their metabolites are found to any substantial extent in the retina [17–19]. In the foveal region of the macula of the primate retina, lutein and zeaxanthin concentrations are at their highest, and they are spatially localized to the Henle fiber layer and to the inner plexiform region [68,21].
The concentration of the macular carotenoids falls precipitously outside of the foveal region so that the concentration of lutein and zeaxanthin in the peripheral retina per unit area is 1% of the concentration at the fovea, and a considerable portion of the extrafoveal carotenoids is associated with the rod outer segments [69–73]. Whenever a tissue exhibits highly selective uptake and deposition of biological molecules, it is likely that specific binding proteins are involved.
In early work from our laboratory, a carotenoid-associated protein from human and bovine retina was identified through photoaffinity labeling and protein microsequencing as tubulin. In vitro, tubulin does exhibit carotenoid-binding properties [71], but with relatively low specificity and affinity [21], and it is likely that it is functioning as a high capacity site for the passive deposition of specifically accumulated carotenoids in much the same way salmon muscle actinin is a site for deposition of high concentrations of astaxanthin [74]. In a follow up study, Crabtree et al. used molecular modeling techniques and presented evidence that carotenoids may be bound to the paclitaxel (Taxol) binding site of the beta-tubulin subunit of microtubules [75].
Subsequently, we continued our pursuit of ocular xanthophyll-binding proteins (XBP) by way of a classical biochemical approach. We maximized the carotenoid-to- protein ratio from the beginning by using 4–8 mm punches of human macula as the starting material and used photodiode array monitoring during chromatography for the nondestructive detection of carotenoid-rich fractions. We homogenized the tissue, performed sucrose gradient centrifugation, solubilized the yellow colored membrane fraction in CHAPS detergent, and then performed ion exchange and gel filtration chromatography. The light yellow XBP fraction that eluted from the final silicagel chromatography step exhibited a characteristic carotenoid absorbance profile, displaying a main absorption maximum at 468 nm that is accompanied with less intense peaks at 438 nm and 488 nm. The absorption spectrum of purified XBP carrying its endogenous carotenoid ligands displayed a bathochromic shift of 9–12 nm as compared to the spectra of lutein and zeaxanthin in organic solvents. XBP’s visible spectrum was identical that that of the macular carotenoid pigment in vivo [21,76].
One-dimensional SDS-PAGE gels of chromatographically purified human macular XBP had only a single major band at ~23 kDa, but two-dimensional (2-D) IEF/SDS-PAGE revealed that there were actually four 21–23 kDa spots with isoelectric points (IEP) ranging from 5.7 to 7.5 when stained with Sypro Ruby. Further analysis by MS/MS fragmentation techniques to obtain additional sequence information using a Mascot search engine and the public NCBI database, allowed us to positively identify the major spot as a pi isoform of human glutathione S-transferase (GSTP1) [21].
GSTP1 is a Phase II detoxification enzyme that is best known to catalyze conjugation of reduced glutathione to toxic compounds targeted for degradation and excretion. It also possesses other unrelated activities including the ability to catalyze double-bond shift reactions in steroid biosynthesis, cis-trans isomerization of retinoic acid, and noncatalytic binding of various hydrophobic molecules “ligandin” activity [77] although carotenoid binding activity had not been described previously [78–79] We therefore used recombinant human GSTP1 to assess its binding properties with xanthophyll carotenoids. We found that GSTP1 binds dietary zeaxanthin and ocular meso-zeaxanthin with high affinity and specificity, while lutein does not bind. Two zeaxanthins were bound per subunit with a Kd of ~0.5 micromolar. Immunocytochemical labeling of primate retina with an antibody against GSTP confirmed that it was localized primarily to the Henle fiber region of the fovea, the region with the highest concentrations of zeaxanthin anywhere in the human body. Functionally, there are two possible roles for the GSTP1/zeaxanthin complex in the primate macula. First, they can act as light screening compounds, efficiently absorbing short wavelength visible light which is considered to be the primary cause of chromatic aberration and the most likely cause of acute and chronic phototoxic damage to the retina. Second, the GSTP1/zeaxanthin complex can enhance zeaxanthin’s antioxidant properties by inhibiting its free radical mediated degradation [80].
GSTP1 is also found in human lenses, and common genetic polymorphisms known to affect GSTP1’s enzymatic functions have been linked to risk of cortical cataracts in certain populations [81]. We are currently repeating these studies to examine whether these same genetic variants play a role in risk of age-related macular degeneration or in determining baseline levels of macular carotenoids. The identity of lutein’s ocular binding protein has proven to be more elusive, and we have therefore turned to another carotenoid-rich tissue, the liver, for further insights.
HEPATIC CAROTENOID BINDING PROTEINS
Along with adipose tissue, liver is a major storage depot for carotenoids in animals, and it is a site for conversion of carotenes to vitamin A. Thus, cellular carotenoid and retinoid binding proteins are expected to be found in abundant amounts in animal liver tissue. Several animal models have been studied in the past to identify carotenoid metabolism and role of hepatic binding proteins [82–83]. Ferrets are known to absorb dietary beta-carotene intact and are considered an ideal animal model for studies on carotenoid absorption and bioavailability. In 1997, Rao et al. reported the existence of a 67 kDa cellular β-carotene-binding protein (CCBP) from ferret liver. Purified CCBP exhibited 460, 482, and 516 nm absorption peaks, indicative of 32 nm bathochromic shifts. This protein was proposed to mediate carotenoid uptake and stabilization in the liver, but it has not been fully purified and sequenced yet [84].
In recent years, the Japanese quail (Coturnix japonica) has become established as another small animal model for studying carotenoid metabolism, especially since its ocular metabolism shares similarities to the human eye [17,85]. High concentrations of lutein, zeaxanthin, and meso-zeaxanthin are present in the quail eye along with a variety of other xanthophylls where they are esterified to fatty acids and stored in photoreceptor oil droplets. Quail liver is also particularly rich in unesterified zeaxanthin and lutein making it an ideal and readily available source for specific xanthophyll binding proteins [17].
We have recently partially purified a lutein-binding protein from liver of Japanese quail (qlLBP). Liver tissue was homogenized using a mechanical homogenizer with buffer containing 50 mM MES, 1 mM EDTA, 20% glycerol, 5 mM CHAPS, 0.5% Triton X-100, 50 μg/ml butylated hydroxytoluene (BHT), and an antiprotease solution (pH 5.5). After homogenization, we proceeded with sucrose density gradient and ultracentrifugation as detailed earlier for ocular xanthophyll-binding proteins [21].
We took advantage of the very high endogenous levels of intensely colored carotenoids in the quail liver as a readily monitored marker for proteins that copurify with carotenoids through multiple biochemical purification steps (Fig 1). We also analyzed carotenoid and protein content at each step of purification (Table 2). Qualitative analysis of carotenoid content revealed that the most purified preparations of the binding protein had lutein as the predominantly associated carotenoid (≥ 90%). Interestingly the bathochromic shift obtained from these preparations was much higher than previously obtained for other LBPs, and is only exceeded by crustacyanin (Fig 1 and Table 1). In our most purified qlLBP preparations, we have only one major band at ~55 kDa, and 2-D gels indicate that it has a pI between 6–7 (Fig 2). We performed binding assays with exogenous lutein using the same protocol we used to study the interactions between GSTP1 and zeaxanthin [18]. Lutein binding to qlLBP was saturable with a dissociation constant of 0.52 micromolar (Fig 3). Mass spectral sequencing is in progress, but it has proven to be quite challenging since protein sequencing databases for quail are poorly developed.
Fig. 1.
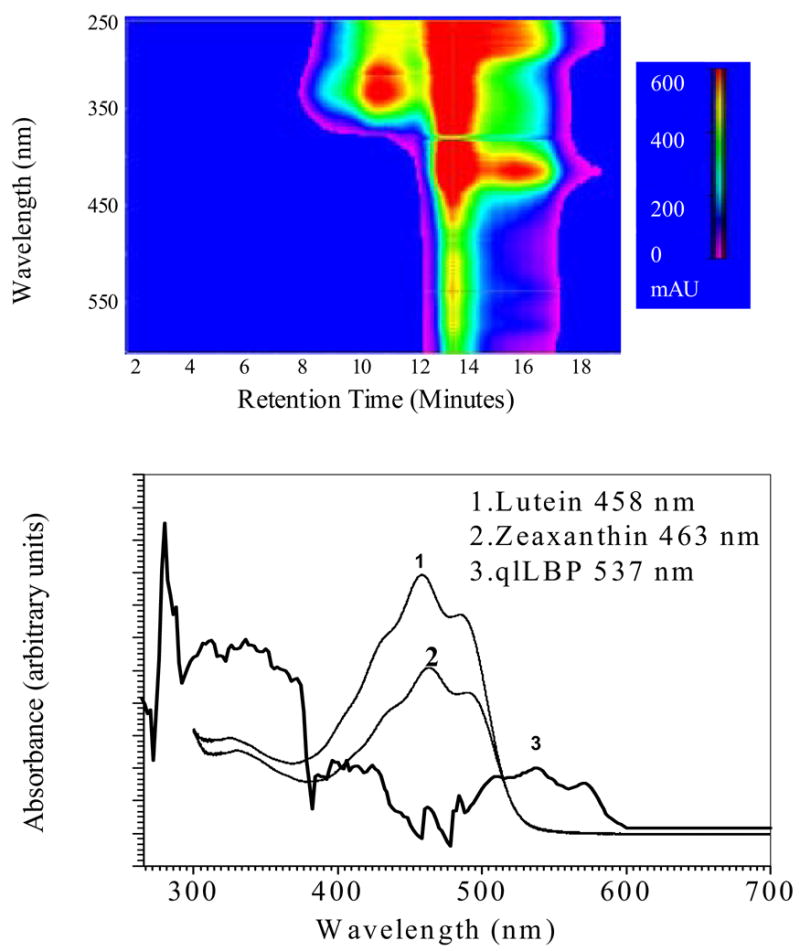
Absorption spectrum of endogenous lutein bound to purified qlLBP. The top figure is the photodiode array chromatogram at the final step of purification on a gel filtration column. In the bottom figure, the absorption spectrum of purified qlLBP at 13 minutes retention time is compared to lutein and zeaxanthin dissolved in elution buffer containing CHAPS detergent.
Table 2.
Purification of quail liver binding protein (qlLBP).
| Steps | Column* | Total Protein (μg/μl) | A537/280 | Lutein (% of total carotenoids) | Purification |
|---|---|---|---|---|---|
| 1 | Extract | 1.4 | 0.02 | 54 | 1 |
| 2 | A (unbound) | 0.84 | 0.039 | 63 | 2 |
| 3 | Q (unbound) | 0.23 | 0.32 | 72 | 16 |
| 4 | S (bound) | 0.08 | 0.62 | 78 | 31.0 |
| 5 | Silica gel | 0.008 | 0.73 | 91 | 36.5 |
A and Q are anion-exchange columns, and S is a cation-exchange column used for purification.
Fig. 2.

A. SDS-PAGE of qlLBP purified from quail liver. 10 μg of purified qlLBP was dissolved in 20 ml of TRIS buffer and loaded on to a 4–20% SDS-polyacrylamide gel for electrophoresis. Later, the gel was stained using Sypro Ruby from Bio-Rad to visualize the protein bands. B. Isoelectric focusing and gel electrophoresis. An aliquot of purified qlLBP was subjected to isoelectric focusing (IEF, pH 3–10) and SDS-PAGE analysis on a 4–20 % gel. Following the run, protein spots were stained with fluorescent Sypro Ruby protein stain. Gels were then visualized using a UV light source.
Fig 3.
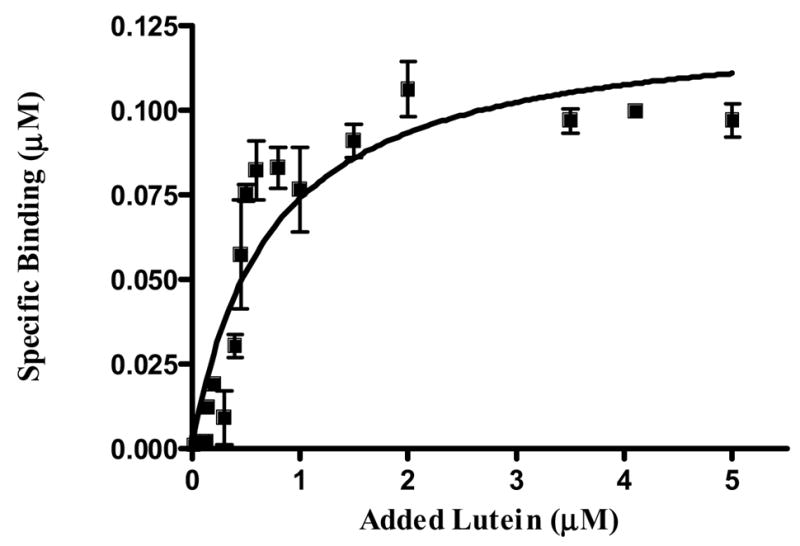
Binding study of partially purified qlLBP with lutein (n=3). Ten μl of concentrated (3R, 3′R, 6′R)-lutein dissolved in THF were added to 490 μl of 50 mM Tris-CHAPS (8 mM) buffer containing 1 μg of protein. After brief mixing, the mixtures were incubated overnight (16 h) at 4 °C. Unbound carotenoids were removed by four cycles of extraction with 200 μl of hexane (Kd=0.52 μM).
Thus, it appears that a novel LBP is present in quail liver, and we have preliminary evidence that a protein with similar spectral properties is present in some vertebrate retinas. Further studies of carotenoid-protein interactions are certain to continue to provide valuable insights into the physiological roles of carotenes and xanthophylls in health and disease in a wide variety of tissues.
Acknowledgments
This work was supported by National Institute of Health Grant EY-11600, the Ruth and Milton Steinbach Fund (New York, NY), and by Research to Prevent Blindness, Inc. (New York, NY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pfander H. Methods in Enzymology. 1992;213:3–13. [Google Scholar]
- 2.Bhosale P. Appl Microbiol Biotechnol. 2004;63:351–361. doi: 10.1007/s00253-003-1441-1. [DOI] [PubMed] [Google Scholar]
- 3.Bhosale P, Bernstein PS. Appl Microbiol Biotechnol. 2005;68:445–455. doi: 10.1007/s00253-005-0032-8. [DOI] [PubMed] [Google Scholar]
- 4.Krinsky NI. Free Radic Biol Med. 1989;7:617–635. doi: 10.1016/0891-5849(89)90143-3. [DOI] [PubMed] [Google Scholar]
- 5.Tee ES. Crit Rev Food Sci Nutr. 1992;31:103–163. doi: 10.1080/10408399209527563. [DOI] [PubMed] [Google Scholar]
- 6.Simpson KL, Chichester CO. Ann Rev Nutr. 1981;1:351–374. doi: 10.1146/annurev.nu.01.070181.002031. [DOI] [PubMed] [Google Scholar]
- 7.Hinds TS, William RD, West L, Knight EM. J Clin Pharmacol. 1997;37:551–558. doi: 10.1002/j.1552-4604.1997.tb04336.x. [DOI] [PubMed] [Google Scholar]
- 8.Edge R, McGarvey DJ, Truscott TG. J Photochem Photobiol B. 1997;41:189–200. doi: 10.1016/s1011-1344(97)00092-4. [DOI] [PubMed] [Google Scholar]
- 9.Foote CS, Denny RW. J Am Chem Soc. 1968;90:6233–6235. [Google Scholar]
- 10.Burton GW, InGold KU. Science. 1984;224:569–573. doi: 10.1126/science.6710156. [DOI] [PubMed] [Google Scholar]
- 11.Hammond BR, Wooten BR, Curran-Celentano J. Arch Biochem Biophys. 1985;385:41–46. doi: 10.1006/abbi.2000.2184. [DOI] [PubMed] [Google Scholar]
- 12.Sies H, Stahl W. Photochem Photobiol Sci. 2004;8:749–752. doi: 10.1039/b316082c. [DOI] [PubMed] [Google Scholar]
- 13.Parker RS. J Nutr. 1989;119:101–104. doi: 10.1093/jn/119.1.101. [DOI] [PubMed] [Google Scholar]
- 14.Deming DM, Teixeira SR, Erdman JW. J Nutr. 2002;132:2700–8. doi: 10.1093/jn/132.9.2700. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin TW. Ann Rev Nutr. 1986;6:273–297. doi: 10.1146/annurev.nu.06.070186.001421. [DOI] [PubMed] [Google Scholar]
- 16.Hata TR, Scholz TA, Ermakov IV, McClane RW, Khachik F, Gellermann W, Pershing LK. J Invest Dermatol. 2000;115:441–448. doi: 10.1046/j.1523-1747.2000.00060.x. [DOI] [PubMed] [Google Scholar]
- 17.Khachik F, de Moura FF, Zhao DY, Aebischer CP, Bernstein PS. Invest Ophthalmol Vis Sci. 2002;43:3383–3392. [PubMed] [Google Scholar]
- 18.Khachik F, Bernstein PS, Garland DL. Invest Ophthalmol Vis Sci. 1997;38:1802–1811. [PubMed] [Google Scholar]
- 19.Bone RA, Landrum JT, Tarsis SL. Vision Res. 1985;25:1531–1535. doi: 10.1016/0042-6989(85)90123-3. [DOI] [PubMed] [Google Scholar]
- 20.Alves-Rodrigues A, Shao A. Toxicol Lett. 2004;150:57–83. doi: 10.1016/j.toxlet.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 21.Bhosale P, Larson AJ, Frederick JM, Southwick K, Thulin CD, Bernstein PS. J Biol Chem. 2004;47:49447–49454. doi: 10.1074/jbc.M405334200. [DOI] [PubMed] [Google Scholar]
- 22.Ruban AV, Young AJ, Horton P. Biochemistry. 1996;35:674678. doi: 10.1021/bi9524878. [DOI] [PubMed] [Google Scholar]
- 23.Bullerjahn GS, Sherman LA. J Bacteriol. 1986;167:396–399. doi: 10.1128/jb.167.1.396-399.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cogdell RJ, Isaacs NW, Freer AA, Arrelano J, Howard TD, Papiz MZ, Hawthornthwaite-Lawless AM, Prince S. Prog Biophys Mol Biol. 1997;68:1–27. doi: 10.1016/s0079-6107(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs NW, Cogdell RJ, Freer AA, Prince SM. Curr Opin Struct Biol. 1995;5:794–797. doi: 10.1016/0959-440x(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 26.Krinsky NI. Biochem Soc Symp. 1995;61:117–126. doi: 10.1042/bss0610117. [DOI] [PubMed] [Google Scholar]
- 27.Reddy KJ, Masamoto K, Sherman DM, Sherman LA. J Bacteriol. 1989;171:3486–3493. doi: 10.1128/jb.171.6.3486-3493.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabunoki H, Sugiyama H, Tanaka Y, Fujii H, Banno Y, Jouni ZE, Kobayashi M, Sato R, Maekawa H, Tsuchida K. J Biol Chem. 1989;277:32133–32140. doi: 10.1074/jbc.M204507200. [DOI] [PubMed] [Google Scholar]
- 29.Jouni ZE, Wells MA. J Biol Chem. 1996;271:14722–14726. doi: 10.1074/jbc.271.25.14722. [DOI] [PubMed] [Google Scholar]
- 30.Zagalsky PF. Acta Crystallogr D Biol Crystallogr. 2003;59:1529–1531. doi: 10.1107/s0907444903013416. [DOI] [PubMed] [Google Scholar]
- 31.Tsuchida K, Arai M, Tanaka Y, Ishihara R, Ryan RO, Maekawa H. Insect Biochem Mol Biol. 1998;28:927–934. doi: 10.1016/s0965-1748(98)00036-8. [DOI] [PubMed] [Google Scholar]
- 32.Gopalapillai R, Kadono-Okuda K, Tsuchida K, Yamamoto K, Nohata J, Ajimura M, Mita K. J Lipid Res. 2006;47:1005–1013. doi: 10.1194/jlr.M500462-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Sakudoh T, Tsuchida K, Kataoka H. Biochem Biophys Res Commun. 2005;336:1125–1135. doi: 10.1016/j.bbrc.2005.08.241. [DOI] [PubMed] [Google Scholar]
- 34.Lazzaro MA, Pepin D, Pescador N, Murphy BD, Vanderhyden BC, Picketts DJ. Mol Endocrinol. 2006 doi: 10.1210/me.2005-0213. In press. [DOI] [PubMed] [Google Scholar]
- 35.Tsuchida K, Jouni ZE, Gardetto J, Kobayashi Y, Tabunoki H, Azuma M, Sugiyama H, Takada N, Maekawa H, Banno Y, Fujii H, Iwano H, Wells MA. J Insect Physiol. 2004;50:363–372. doi: 10.1016/j.jinsphys.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Tabunoki H, Higurashi S, Ninagi O, Fujii H, Banno Y, Nozaki M, Kitajima M, Miura N, Atsumi S, Tsuchida K, Maekawa H, Sato RA. FEBS Lett. 2004;567:175–178. doi: 10.1016/j.febslet.2004.04.067. [DOI] [PubMed] [Google Scholar]
- 37.Britton G, Weesie RJ, Askin D, Warburton JD, Gallardo-Guerrero L, Jansen FJ, de Groot HJM, Lugtenburg J, Cornard JP, Merlin JC. Pure Appl Chem. 1997;69:2075–2084. [Google Scholar]
- 38.Zagalsky PF, Eliopoulos EE, Findlay JB. Biochem J. 1991;274:79–83. doi: 10.1042/bj2740079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlikowski M. Acta Phys Pol. 1988;74:145–149. [Google Scholar]
- 40.Buchwald M, Jencks WP. Biochemistry. 1968;7:844–859. doi: 10.1021/bi00842a043. [DOI] [PubMed] [Google Scholar]
- 41.Van Wijk AA, Spaans A, Uzunbajakava N, Otto C, de Groot HJ, Lugtenburg J, Buda F. J Am Chem Soc. 2005;127:1438–1445. doi: 10.1021/ja045049+. [DOI] [PubMed] [Google Scholar]
- 42.Zagalsky PF. Methods Enzymol. 1985;111B:216–247. doi: 10.1016/s0076-6879(85)11011-6. [DOI] [PubMed] [Google Scholar]
- 43.Cianci M, Rizkallah PJ, Olczak A, Raftery J, Chayen NE, Zagalsky PF, Helliwell JR. Acta Crystallogr D Biol Crystallogr. 2001;57:1219–1229. doi: 10.1107/s0907444901009350. [DOI] [PubMed] [Google Scholar]
- 44.Cianci M, Rizkallah PJ, Olczak A, Raftery J, Chayen NE, Zagalsky PF. Proc Natl Acad Sci. 2002;99:9795–9800. doi: 10.1073/pnas.152088999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein PS, Yoshida MD, Katz NB, McClane RW, Gellermann W. Invest Ophthalmol Vis Sci. 1998 ;39:2003–2011. [PubMed] [Google Scholar]
- 46.Brush AH. FASEB J. 1990;12:2969–2977. doi: 10.1096/fasebj.4.12.2394316. [DOI] [PubMed] [Google Scholar]
- 47.Lakshman MR. J Nutr. 2004;134:241S–245S. doi: 10.1093/jn/134.1.241S. [DOI] [PubMed] [Google Scholar]
- 48.Olson JA, Hayaishi O. Proc Natl Acad Sci U S A. 1965;54:1364–1370. doi: 10.1073/pnas.54.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman DS, Huang HS. 1965;149:879–880. doi: 10.1126/science.149.3686.879. [DOI] [PubMed] [Google Scholar]
- 50.von Lintig J, Hessel S, Isken A, Kiefer C, Lampert LM, Voolstra O, Vogt K. Biochim Biophys Acta. 2005;1740:122–131. doi: 10.1016/j.bbadis.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 51.von Lintig J, Vogt K. J Nutr. 2004;134:251S–2516S. doi: 10.1093/jn/134.1.251S. [DOI] [PubMed] [Google Scholar]
- 52.Wyss A, Wirtz G, Woggon W, Brugger R, Wyss M, Friedlein H, Bachmann H, Hunziker W. Biochem Biophys Res Commun. 2000;271:334–336. doi: 10.1006/bbrc.2000.2619. [DOI] [PubMed] [Google Scholar]
- 53.Von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K. Proc Natl Acad Sci. 2001;98:1130–1135. doi: 10.1073/pnas.031576398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redmond TM, Gentleman S, Duncan T, Yu S, Wiggert B, Gantt E, Cunningham FX. J Biol Chem. 2001;276:6560–6565. doi: 10.1074/jbc.M009030200. [DOI] [PubMed] [Google Scholar]
- 55.Yan W, Jang GF, Haeseleer F, Esumi N, Chang J, Kerrigan M, Campochiaro M, Campochiaro P, Palczewski K, Zack DJ. Genomics. 2001;72:193–202. doi: 10.1006/geno.2000.6476. [DOI] [PubMed] [Google Scholar]
- 56.Trudel E, Beaufils S, Renault A, Breton R, Salesse C. Biochemistry. 2006;45:3337–3347. doi: 10.1021/bi0519405. [DOI] [PubMed] [Google Scholar]
- 57.Lindqvist A, He YG, Andersson S. J Histochem Cytochem. 2005;53:1403–1412. doi: 10.1369/jhc.5A6705.2005. [DOI] [PubMed] [Google Scholar]
- 58.Krinsky NI, Cronwell DG, Oncley JL. Arch Biochem Biophys. 1958;73:233–246. doi: 10.1016/0003-9861(58)90259-5. [DOI] [PubMed] [Google Scholar]
- 59.Khachik F, Beecher GR, Goli MB, Lusby WR, Daitch CE. Methods Enzymol. 1992;213:205–219. doi: 10.1016/0076-6879(92)13122-e. [DOI] [PubMed] [Google Scholar]
- 60.Khachik F, Beecher GR, Smith JC., Jr J Cell Biochem. 1995;22:236–246. doi: 10.1002/jcb.240590830. [DOI] [PubMed] [Google Scholar]
- 61.Boileau AC, Merchen NR, Wasson K, Atkinson CA, Erdman JW. J Nutr. 1999;129:1176–1181. doi: 10.1093/jn/129.6.1176. [DOI] [PubMed] [Google Scholar]
- 62.Greene CM, Waters D, Clark RM, Contois JH, Fernandez ML. Nutr Metab (Lond) 2006;3:6. doi: 10.1186/1743-7075-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohashi M, Carr BR, Simpson ER. Endocrinology. 1982;110:1477–1482. doi: 10.1210/endo-110-5-1477. [DOI] [PubMed] [Google Scholar]
- 64.Kiefer C, Sumser E, Wernet MF, Von Lintig J. Proc Natl Acad Sci. 2002;99:10581–10586. doi: 10.1073/pnas.162182899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Southon S. Nutr Metab Cardiovasc Dis. 2001;4:78–81. [PubMed] [Google Scholar]
- 66.Zsila F, Simonyi M, Lockwood SF. Bioorg Med Chem Lett. 2003;13:4093–4100. doi: 10.1016/j.bmcl.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 67.Zsila F, Bikadi Z, Simonyi M. Biochem Pharmacol. 2002;64:1651–1660. doi: 10.1016/s0006-2952(02)01350-3. [DOI] [PubMed] [Google Scholar]
- 68.Snodderly DM, Auran JD, Delori FC. Invest Ophthalmol Vis Sci. 1984;25:674–685. [PubMed] [Google Scholar]
- 69.Bone RA, Landrum JT, Fernandez L, Tarsis SL. Invest Ophthalmol Vis Sci. 1988;29:843–849. [PubMed] [Google Scholar]
- 70.Handelman GJ, Dratz EA, Reay CC, van Kuijk JG. Invest Ophthalmol Vis Sci. 1988;29:850–855. [PubMed] [Google Scholar]
- 71.Bernstein PS, Balashov NA, Tsong ED, Rando RR. Invest Ophthalmol Vis Sci. 1997;38:167–175. [PubMed] [Google Scholar]
- 72.Rapp LM, Maple SS, Choi JH. Invest Ophthalmol Vis Sci. 2000;41:1200–1209. [PubMed] [Google Scholar]
- 73.Sommerburg OG, Siems WG, Hurst JS, Lewis JW, Kliger DS, van Kuijk FJ. Curr Eye Res. 1999;19:491–495. doi: 10.1076/ceyr.19.6.491.5276. [DOI] [PubMed] [Google Scholar]
- 74.Matthews SJ, Ross NW, Lall SP, Gill TA. Comp Biochem Physiol B, Biochem Mol Biol. 2006;144:206–214. doi: 10.1016/j.cbpb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 75.Crabtree DV, Ojima I, Geng X, Adler AJ. Bioorg Med Chem. 2001;9:1967–1976. doi: 10.1016/s0968-0896(01)00103-1. [DOI] [PubMed] [Google Scholar]
- 76.Yemelyanov AY, Katz NB, Bernstein PS. Exp Eye Res. 2001;72:381–392. doi: 10.1006/exer.2000.0965. [DOI] [PubMed] [Google Scholar]
- 77.Habig WH, Pabst MJ, Fleischner G, Gatmaitan Z, Arias IM, Jakoby WB. Proc Natl Acad Sci. 1974;71:3879–3882. doi: 10.1073/pnas.71.10.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lederer B, Boger P. Biochim Biophys Acta. 2003;1621:226–233. doi: 10.1016/s0304-4165(03)00073-4. [DOI] [PubMed] [Google Scholar]
- 79.Chen H, Juchau MR. Biochem J. 1997;327:721–726. doi: 10.1042/bj3270721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhosale P, Bernstein PS. Biochim Biophys Acta. 2005;1740:116–121. doi: 10.1016/j.bbadis.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Juronen E, Tasa G, Veromann S, Parts L, Tiidla A, Pulges R, Panov A, Soovere L, Koka K, Mikelsaar AV. Investig Ophthalmol Vis Sci. 2000;41:2262–2267. [PubMed] [Google Scholar]
- 82.Ong DE, Chytil F. Proc Natl Acad Sci. 1976;73:3976–3978. doi: 10.1073/pnas.73.11.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee CM, Boileau AC, Boileau TW, Williams AW, Swanson KS, Heintz KA, Erdman JW. J Nutr. 1999;129:2271–2277. doi: 10.1093/jn/129.12.2271. [DOI] [PubMed] [Google Scholar]
- 84.Rao MN, Ghosh P, Lakshman MR. J Biol Chem. 1997;272:24455–24460. doi: 10.1074/jbc.272.39.24455. [DOI] [PubMed] [Google Scholar]
- 85.Toyoda Y, Thomson LR, Langner A, Craft NE, Garnett KM, Nichols CR, Cheng KM, Dorey CK. Invest Ophthalmol Vis Sci. 2002;43:1210–1221. [PubMed] [Google Scholar]


