Abstract
Adenosine (A) to inosine (I) RNA editing occurs widely in the human transcriptome, and a large proportion of editing sites are within untranslated regions (UTRs). MicroRNAs (miRNAs), an abundant class of regulatory genes, specify the expression of a large number of target genes by pairing to their 3′ UTRs. To study the interplay between these two post-transcriptional events, we developed a computational pipeline to integrate sequence and miRNA tissue specificity data. The results show that some A-to-I RNA editing positions have a potential to block the miRNA:target recognition, although further computational simulation suggests that RNA editing tends to avoid miRNA target sites in general. We propose that a small proportion of RNA editing events may provide an additional layer of control on miRNA-mediated repression. Further investigation is needed to elucidate the functional effect of these special RNA editing events.
Keywords: A-to-I RNA editing, microRNA repression, microRNA target sites, tissue specificity
INTRODUCTION
RNA editing generates RNA diversity through the post-transcriptional modification of single nucleotides in pre-mRNA (Maas et al. 2003). Adenosine (A) to inosine (I) modification by adenosine deaminases (ADARs), with inosine acting as guanosine (G) during translation, appears to be the most common type of nuclear RNA editing in eukaryotes (Bass 2002). Several recent studies have reported that A-to-I editing occurs widely in the human transcriptome (at least 2% of publicly available mRNAs), and most editing sites reside in Alu repetitive elements, which are typically 300 nucleotides (nt) long and comprise >10% of the human genome (Athanasiadis et al. 2004; Kim et al. 2004; Levanon et al. 2004, 2005; Eisenberg et al. 2005). Because the vast majority of edited nucleotides are within intronic or untranslated regions (UTRs), the functional consequence of RNA editing is often hard to infer. To date, except for a few well-studied cases (Higuchi et al. 1993; Burns et al. 1997; Rueter et al. 1999), the biological role of much A-to-I editing is still under debate.
MicroRNAs (miRNAs) are endogenous ∼22-nt noncoding RNAs that can play an important role in the regulation of gene expression by pairing to the messages of protein-coding genes, thereby specifying mRNA cleavage or repression of productive translation (Ambros 2004; Bartel 2004; Zamore and Haley 2005). In vertebrates, miRNA genes are one of the most abundant classes of regulatory genes (∼1% of all the genes) (Lim et al. 2003, 2005; Bartel 2004; Bartel and Chen 2004). Recent studies have indicated that target recognition by one miRNA is mainly achieved by pairing to the nucleotides at positions 2–7 of the miRNA (the so-called “miRNA seed”), and often a single mutation in the miRNA seed match of the target site can lead to total abolishment of effective repression (Lewis et al. 2003; Bartel 2004; Brennecke et al. 2005; Stark et al. 2005). In fact, several computational methods have been developed to predict miRNA target genes based on this principle, suggesting that 20%–30% of protein-coding genes are under selective pressure to contain conserved 7-nt matches to miRNA seed regions, and that these regions are likely to be miRNA targets (Lewis et al. 2003, 2005; Rajewsky 2006). Very recently, a study further suggested that nonconserved 7-nt matches are also able to mediate repression when exposed to the miRNA, implying a widespread impact of miRNAs on gene expression (Farh et al. 2005). (For simplicity, throughout the rest of this paper we often refer to these 7-nt matches as miRNA target sites, recognizing that they are necessarily biological target sites.)
Recently, A-to-I editing of miRNA precursor molecules has been reported (Luciano et al. 2004; Blow et al. 2006); however, the potential role of the editing on miRNA target genes remains unexplored. Given the wide occurrence of both A-to-I editing and miRNA target sites in 3′-UTR regions, we wonder whether some editing events can interrupt (or create) functional target sites, thus adding another layer of control to miRNA-mediated repression. Therefore, by integrating sequence and miRNA tissue specificity data, we developed a computational pipeline to predict miRNA target sites subject to RNA editing. Intriguingly, we found that some edited positions lie in putative miRNA target sites. Our work calls attention to the layers of complexity that govern human gene regulation, and more experimental work is needed to further elucidate the functional effect of these RNA editing events.
RESULTS
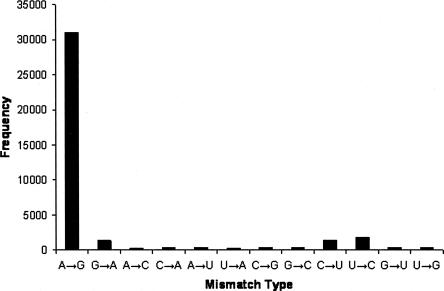
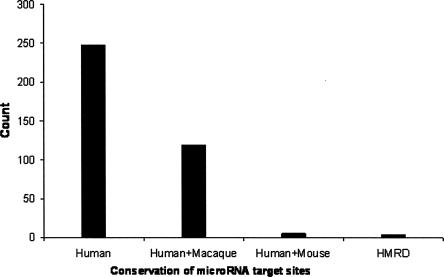
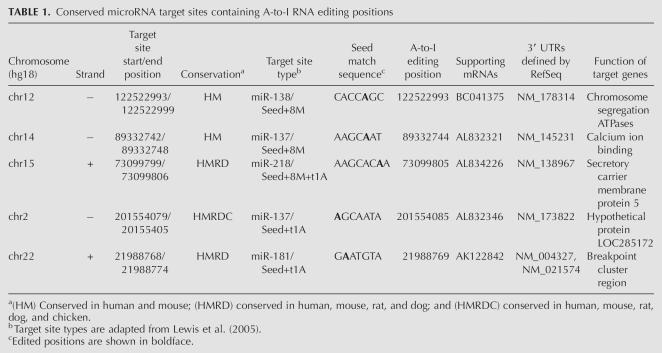
We first identified a set of highly specific A-to-I editing sites in the human genome, using the edited transcripts from two previous large-scale studies (Athanasiadis et al. 2004; Kim et al. 2004). As shown in Figure 1, there is a striking excess of A-to-G mismatches in these transcripts when aligned with the reference human genome. If we assume that the noise (such as sequencing error and single nucleotide polymorphism) in our data set is roughly equal to the number of G-to-A mismatches, the accuracy of A-to-I editing positions is ∼96% (1349/31,014 ≈ 4%). Based on the annotation of RefSeq genes, 3236 nonredundant editing positions are found in 3′ UTRs. Among them, we found that 292 editing positions reside in a miRNA's 7-nt target site, which includes a 6-nt match to the miRNA seed (positions 2–7) and either a seventh Watson–Crick match to position 8 of the miRNA (designated as the m8 position) or an adenosine at position 1 of the target site (designated as the t1 position; Supplemental Data file 1). To evaluate the biological significance of the occurrence of these special editing sites, we developed a “local clustering and sampling” algorithm to simulate the random occurrence of RNA editing events in 3′ UTRs (see Materials and Methods). We found the observed number of editing positions interrupting miRNA target sites to be significantly smaller than random expectation (z score = 5.0, P value < 6 × 10−7; Supplemental Data file 2), suggesting that RNA editing in 3′ UTRs may be under functional constraint to avoid interrupting miRNA target sites. We further studied the conservation of these interrupted miRNA target sites (Fig. 2). Because A-to-I editing is predominantly associated with primate-specific Alu elements, we found that most target sites are not conserved beyond primates, but we did find a handful of target sites conserved in mouse, rat, and dog (Table 1).
FIGURE 1.
The frequency of various mismatch types in A-to-I edited transcripts.
FIGURE 2.
Conservation of microRNA target sites interrupted by RNA editing. (HMRD) Conserved in human, mouse, rat, and dog.
TABLE 1.
Conserved microRNA target sites containing A-to-I RNA editing positions
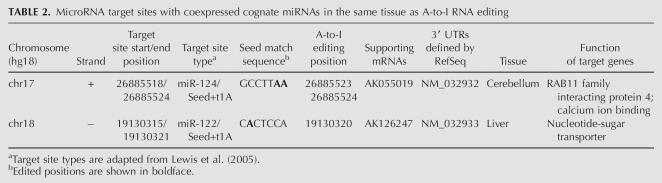
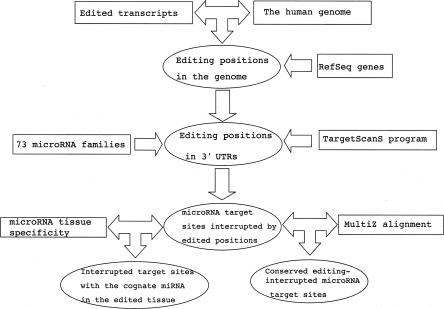
Since a recent study has suggested that a 7-nt target site (no matter whether it is conserved or not) is often sufficient to mediate suppression when exposed to the cognate miRNA (Farh et al. 2005), we further incorporated miRNA tissue specificity data to identify the editing-interrupted miRNA target sites whose cognate miRNA (family) is coexpressed in the same tissue in which RNA editing occurs. As a result, we found two target sites interrupted by three editing positions (Table 2). The pipeline for our computational analysis is shown in Figure 3.
TABLE 2.
MicroRNA target sites with coexpressed cognate miRNAs in the same tissue as A-to-I RNA editing
FIGURE 3.
The computational pipeline to identify microRNA target sites subject to RNA editing.
In addition, our study also revealed that ∼200 A-to-I editing positions have a potential to create a new miRNA target site, if we assume that inosine works as well as guanine in miRNA:target recognition (Supplemental Data file 3).
DISCUSSION
Based on our sequence analysis, we first identified a set of A-to-I editing positions that can interrupt 7-nt matches to miRNA seed regions. Most of them should have no functional effect because (1) the 7-nt matches either are not real miRNA target sites or (2) they are miRNA target sites but function in different tissues relative to RNA editing. However, we cannot rule out the possibility that a few RNA editing positions may interrupt functional miRNA target sites and thus block miRNA:target recognition. To elucidate this dual-level regulation, it will be necessary to verify the miRNA target sites and evaluate the functional effect of the specific RNA editing within them. In particular, two types of miRNA target sites that we identified seem most appealing to examine: (1) the interrupted miRNA target sites that have been conserved in the long history of eutherian evolution; and (2) the target sites whose cognate miRNA is coexpressed in the tissue where RNA editing takes place.
Undoubtedly, the possible RNA editing of miRNA target sites that this study reveals is only the tip of the iceberg. First, we note that only a small proportion of the human transcriptome is available so far. Second, only those transcripts with extensive A-to-I editing can be confidently identified by the computational and statistical methods. Third, due to the incomplete annotation of transcripts in the public database and the limitation of miRNA tissue specificity data, only a small number of tissues could presently be investigated.
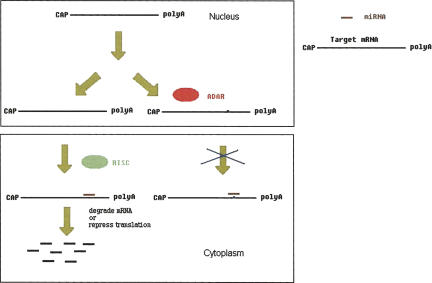
If some RNA editing events really interrupt functional miRNA target sites, they may represent some very specific cases of coordinated regulation. One possible scenario (Fig. 4) is that two mRNA versions of a gene might be produced in the nucleus, and in the cytoplasm the unedited transcripts are subject to miRNA repression, while the miRNA binding to edited transcripts is blocked. Since the editing efficiency usually ranges from <2% to 90% at individual positions (Athanasiadis et al. 2004), the editing may add a finer control to miRNA-mediated repression and thus influence the final protein products in a “quantitative” rather than “qualitative” manner.
FIGURE 4.
A model of the interplay between RNA editing and microRNA-mediated repression.
MATERIALS AND METHODS
To identify RNA editing positions relevant to miRNA target sites, we first compiled a set of mRNA transcripts under A-to-I editing from two recent large-scale studies that used both statistical and experimental approaches to identify edited transcripts (Athanasiadis et al. 2004; Kim et al. 2004). Second, we downloaded the mRNA sequences and their genomic locations from the UCSC genome browser and aligned the mRNAs with the reference human genome (hg18). Then, RNA editing positions were inferred from the A-to-G discrepancies between the genomic and mRNA sequences. As a result, based on 2454 edited mRNAs, our data set includes 28,311 nonredundant A-to-I RNA editing positions. Next, we identified the editing positions within the 3′-UTR regions that were defined by well-annotated RefSeq genes. Finally, for each of these editing positions, we extracted its local genomic and mRNA sequences and evaluated whether the RNA editing has a potential to interrupt (or create) a miRNA target site (a 7- or 8-nt match to a miRNA seed region) using the TargetScanS program (Lewis et al. 2005). For this purpose, we used the same set of 73 conserved miRNA families as in Farh et al. (2005) and obtained the miRNA sequences from the miRBase sequence database (Griffiths-Jones 2004).
To evaluate the biological significance of RNA editing positions interrupting a potential miRNA target site, we developed a local clustering and sampling (LCS) algorithm to simulate the random occurrence of RNA editing in 3′ UTRs. Because RNA editing positions tend to be clustered in the sequence, our LCS algorithm maintained this clustering feature in the randomization, instead of treating each position independently. First, we defined two RNA editing positions as neighbors if they were within a small distance (10 nt), and then we used the single-linkage algorithm to group editing positions into clusters. That is, if editing positions A and B are neighbors and positions B and C are neighbors, we group editing positions A, B, and C together into a cluster, regardless of whether or not positions A and C are neighbors. This process assigns each editing position to one cluster only. Second, for each cluster of editing positions, we defined a sampling window, which includes the region from the editing position at the 5′ end to the one at the 3′ end in the cluster plus five flanking nucleotides at each side. Third, for each editing cluster, given the number of editing positions within the cluster, we randomly chose the same number of adenosine positions from the sampling window as pseudoediting positions. Then, given the pseudoediting positions and the presence of miRNA target sites (7-mer seed matches regardless of whether conserved or not) in 3′ UTRs, we calculated the total number of positions interrupting a miRNA target site over all the clusters. This step was repeated 1000 times to generate an empirical background distribution (the number of editing positions interrupting a miRNA target site versus frequency). Based on this distribution, we derived the probability of the number of miRNA-target-interrupting editing positions observed in the real data set. By using this algorithm, we not only controlled the number of RNA editing positions in each 3′ UTR in the randomization, but also maintained (1) the spatial clustering of editing positions, (2) the local nucleotide composition (e.g., GC%), and (3) the relative location of editing positions in 3′ UTRs. Moreover, the results remained essentially the same when different cutoff values were explored.
To study the conservation of miRNA target sites interrupted by RNA editing, we further extracted the corresponding orthologous 3′-UTR regions (human [hg18], macaque [rheMac2], mouse [mm8], rat [rn4], dog [canFam2], and chicken [galGal2]) from the UCSC MultiZ17 alignments (Blanchette et al. 2004).
To study the mRNA targets with coexpressed cognate miRNAs, we estimated the tissue specificity of the miRNA families using the data from Farh et al. (2005), where the signal (as probabilities) of the 73 miRNA families in 61 mouse tissues was inferred based on “target-site depletion.” For our analysis, the miRNA tissue specificity is assumed to be the same in the human homologous tissues. Given the probabilities and a false discovery rate of 10%, we chose 0.0015 as the threshold to label a family of miRNA genes as “expressed in a given tissue.”
SUPPLEMENTAL DATA
Supplemental Data files can be found at http://oxytricha.princeton.edu/liang/editing/editing.htm.
ACKNOWLEDGMENTS
We thank David Bartel (MIT) and two anonymous reviewers for their valuable suggestions. We also thank laboratory members Joey Zhou, Tom Doak, and Mariusz Nowacki for discussions. This work was supported by National Institute of General Medical Sciences grant GM59708 to L.F.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.296407.
REFERENCES
- Ambros, V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Athanasiadis, A., Rich, A., Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P., Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F., Roskin, K.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow, M.J., Grocock, R.J., van Dongen, S., Enright, A.J., Dicks, E., Futreal, P.A., Wooster, R., Stratton, M.R. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Stark, A., Russell, R.B., Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg, E., Nemzer, S., Kinar, Y., Sorek, R., Rechavi, G., Levanon, E.Y. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Farh, K.K., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., Bartel, D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., Seeburg, P.H. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Kim, D.D., Kim, T.T., Walsh, T., Kobayashi, Y., Matise, T.C., Buyske, S., Gabriel, A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon, E.Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z.Y., Shoshan, A., Pollock, S.R., Sztybel, D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Levanon, K., Eisenberg, E., Rechavi, G., Levanon, E.Y. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep. 2005;6:831–835. doi: 10.1038/sj.embor.7400507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., Bartel, D.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Luciano, D.J., Mirsky, H., Vendetti, N.J., Maas, S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas, S., Rich, A., Nishikura, K. A-to-I RNA editing: Recent news and residual mysteries. J. Biol. Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- Rajewsky, N. microRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Rueter, S.M., Dawson, T.R., Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Bushati, N., Russell, R.B., Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′-UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Haley, B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]