Abstract
Most of the vertebrate genome is transcribed into RNA. Transcribed regions contain hundreds of thousands of potential duplex structures that could serve as substrates for RNAse III enzymes of microRNA (miRNA) maturation pathways. Yet, only a minority of these potential precursors make their way to the cytoplasm to form mature miRNAs. We question here what specific structural features make an RNA stem–loop structure an adequate primary or precursor miRNA. We address this question by comparing known pre-miRNAs to other predicted noncoding transcripts obtained from comparative genomics scans, using the structure comparison program RNAforester. By analyzing a classification tree of 1200 such RNA structures, we observe that pre-miRNAs cluster distinctly from other duplex structures of apparently similar size and free energy. The most distinctive features of nonprecursor duplexes are increased lengths and numbers of bulges and internal loops when compared to real miRNA precursors. Thanks to these characteristics, secondary structure comparison can predict the miRNA precursor status of a candidate stem–loop with a surprising accuracy. Furthermore, predicted noncoding transcripts tend to depart from miRNA precursor characteristics more strongly than randomly occurring duplex structures in genomic DNA. This result suggests that many noncoding RNAs may be under selection to dodge the RNAi pathway.
Keywords: Dicer, Drosha, RNAse III, miRNA precursor, secondary structure
INTRODUCTION
Mature microRNAs (miRNAs) are 21- to 23-nucleotide (nt)-long, single-stranded RNAs processed from longer transcripts by two RNAse III-like enzymes, Drosha and Dicer. Drosha acts in the nucleus on polycistronic primary miRNAs (pri-miRNA), to produce a 70-nt pre-miRNA. Dicer acts in the cytoplasm on the pre-miRNA to produce the mature miRNA. The distinctive component of pri- and pre-miRNA is a long duplex containing several bulges or mispairs, capped by an apical loop of variable size. Little is known to date about the structural features that make such a duplex an adequate substrate for Drosha or Dicer. Mutagenesis of miR-30 and miR-21 precursors (Zeng and Cullen 2003) showed that mutations in the primary sequence have little effect on miRNA expression as long as a certain level of base-pairing is maintained, base-pair formation at the base of the duplex being particularly important. Recent X-ray structure of the uncomplexed Dicer (MacRae et al. 2006) showed that Dicer may recognize a stretch of duplex RNA equivalent to 25 base pairs (bp), but did not identify more specific topological constraints on the RNA ligand. According to Lai et al. (2003), a minimal miRNA precursor stem is ∼23 bp long and includes at most three mismatches. However, miRNA databases contain shorter precursors with only 15 bp in the stem (Griffiths-Jones 2006). Requirements for Drosha processing include a longer duplex of ∼33 bp, as well as the presence of essential unpaired basal segments (Han et al. 2006).
Long duplexes are frequent in noncoding RNAs (ncRNAs) and other transcribed sequences. There are three 33-bp stems—allowing for four mispairs and one 2-nt bulge—and seventeen 23-bp stems in the highly expressed 18S and 28S rRNAs. A search for potential 33-bp stems in human genomic DNA finds about one hit every 10 kb (Supplemental Table 1). Considering that most of the mammalian genomic DNA is transcribed (Cheng et al. 2005), one can be troubled by the idea of vast amounts of duplex-forming RNAs encountering RNAse III enzymes in the nucleus first, and in the cytoplasm if exported. This eventuality raises the fundamental question of miRNA identity. How do Dicer and Drosha discriminate their legitimate substrates from other duplex structures that can arise serendipitously in other transcripts? Besides its direct biological interest, this question relates to our ability to distinguish bona fide precursors or primary miRNAs from other conserved genomic sequences. Indeed, most computational miRNA discovery methods involve folding conserved genomic elements and filtering candidates in function of their secondary structure (Lewis et al. 2003; Wang et al. 2004). As folding free energy or stem length alone is not sufficient to discriminate miRNA precursors from other structures, additional criteria have been considered, including preference for a 5′ Uracil (Lim et al. 2003), stem or bulge symmetry (Lim et al. 2003; Pfeffer et al. 2005), and frequencies of 3-nt elements combining base-pairing and sequence characters (Xue et al. 2005). Although these parameters have indeed improved miRNA prediction, a posterior analysis of the most distinctive topological features that make a predicted duplex structure an adequate substrate for RNase III has not been carried out to date.
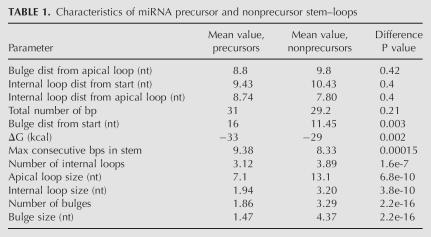
TABLE 1.
Characteristics of miRNA precursor and nonprecursor stem–loops
LARGE-SCALE CLUSTERING OF ncRNA SECONDARY STRUCTURES
To examine topological features in RNA stem–loops, we used the secondary structure alignment program RNAforester (Höchsmann et al. 2003, 2004), which performs a structure comparison using a tree alignment algorithm and computes a distance measure that depends on secondary structure shapes rather than primary sequence. The local comparison option of RNAforester was used in order to identify stem–loops contained in larger RNA sequences. We questioned the ability of such a method to discriminate miRNAs from other RNAs in a large pan-genome collection of ncRNA. This collection was provided by the works of Washietl et al. (2005), who evaluated the folding potential of each conserved element in the human genome based on energy and base-pair covariation, producing a list of 30,000 putative RNAs. We extracted from this list the subset of 1200 putative human ncRNAs that were nonoverlapping and not part of multigenic families or repeats. This collection contained 78 miRNAs and 51 other known ncRNAs (Supplemental Table 2).
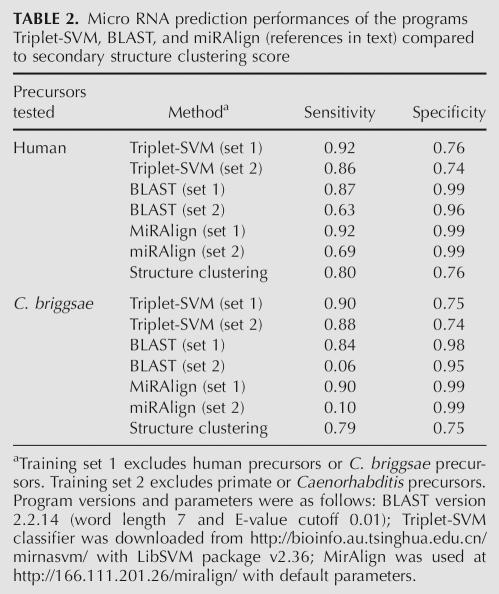
TABLE 2.
Micro RNA prediction performances of the programs Triplet-SVM, BLAST, and miRAlign (references in text) compared to secondary structure clustering score
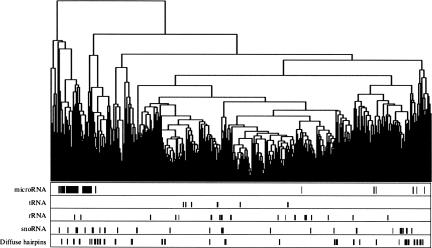
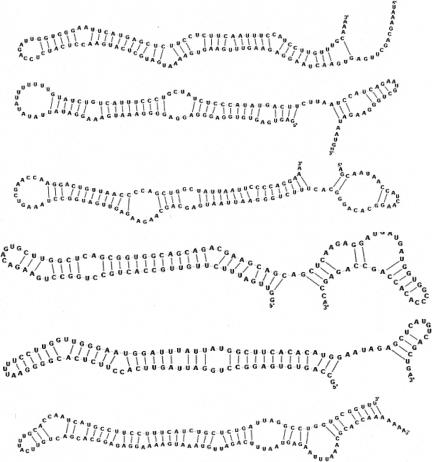
Through a pairwise RNAforester comparison of the 1200 structures, we produced a hierarchical clustering tree, shown in Figure 1 with locations of known ncRNAs. Most miRNAs were grouped in a single cluster, whereas other ncRNAs (tRNA, rRNA, snoRNA) were more diffuse, possibly because more complex structures are more likely to be predicted only in part by the initial computational pipeline, and such partial structures are expected to fail the comparison tests more often. Although the structure-based comparison performed relatively well on miRNA precursors, we were surprised that many other stem–loop structures that were unknown to the Rfam registry were dispersed through the tree while not obviously different from the miRNAs precursors. The precursor structures in mirBase (version 8; Griffiths-Jones 2006) display a minimum length of 15 bp and a minimum free energy of −20 kcal. Sixty-seven structures in our tree correspond to these criteria and yet are not annotated in mirBase and do not cluster with any known miRNAs precursors (Fig. 1). The average free energy and size of the main duplex for all diffuse hairpins were, respectively, −29 Kcal and 29 bp, versus −35 Kcal and 32 bp for miRbase precursors. A selection of such “diffuse hairpins” with apparent highly stable structures is shown in Figure 2.
FIGURE 1.
Clustering of conserved RNA secondary structures. Candidate human ncRNA sequences were obtained from the RNAz screen of Washietl et al. (2005). Twelve hundred ncRNA candidates were obtained by removing overlapping sequences and sequences with inconsistent genome coordinates, and retaining a single sequence per paralogous set. Sequences were folded using RNAalifold (Hofacker et al. 2001) and then compared pairwise using RNAforester (Höchsmann et al. 2004) using normalized scores for each comparison. RNAforester options were local comparison and no use of sequence information (no Ribosum). The resulting 1200×1200/2 matrix was then used to create an UPGMA cluster using mean values. Tick marks at bottom indicate position of known RNA genes and of non-miRNA hairpin structures (diffuse hairpins).
FIGURE 2.
Sample of conserved human stem–loop structures that do not cluster with miRNA precursors. Structures were drawn using the jViz.Rna program (http://jviz.research.iat.sfu.ca).
COCLUSTERING WITH miRNA PRECURSORS
In order to measure the resemblance of any given stem–loop to a miRNA precursor, we devised a metrics that evaluates the ability of a sequence to cluster with miRNA precursors and not with diffuse hairpins. We assumed diffuse hairpins could be an appropriate negative control as these sequences are conserved and predicted to be expressed as ncRNAs and therefore should harbor structural features preventing them from entering the RNAi pathway. A candidate sequence is compared to each of the sequences in the miRNA-enriched clade (109 sequences) and each of the diffuse hairpin sequences (67 sequences) using RNAforester. In order to reduce the false-negative rates, we considered possible alternative structures for the candidate sequence using the RNAshapes program (Steffen et al. 2006), which analyzes the different structural possibilities within a given energy range and displays structures that show different spatial conformations. Each of the structures given by RNAshapes is considered separately, and the highest scoring candidate is retained. All pairwise comparisons are then classified from best to worst RNAforester score. For each of the top 20 scores, a number of points is given to the candidate. If the high scoring sequence is one of the diffuse hairpins, zero points are given. If the sequence comes from the miRNA-enriched cluster, a number of points is given to the candidate in function of the sequence's rank (20 if the sequence had the best RNAforester score, 19 if it was second best, etc.). This is a simple way to give more weight to higher scoring pairwise comparisons. The final score is the sum of these points brought down to a zero-to-100 scale, 100 being a good candidate and zero a poor candidate.
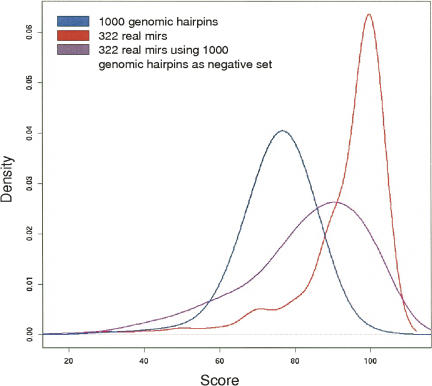
Figure 3 shows score distributions obtained with different sets of structures. The blue curve shows scores of 1000 random human genome fragments that are predicted to form hairpin structures with energy/length criteria <−20 Kcal mol−1/15 bp, which would have triggered a positive hairpin candidate by other studies (Wang et al. 2004; Bentwich et al. 2005; Cui et al. 2006). The red curve shows scores obtained by 322 known miRNAs from miRbase. These two curves are well separated, indicating that the computationally predicted secondary structure is, in itself, an important miRNA identity element. We changed the scoring procedure so that the negative control set was made of the 1000 randomly selected hairpin structures instead of the 67 diffuse hairpins. Interestingly, the resulting score distribution for real miRNAs (Figure 3, purple curve) strongly overlaps that of random hairpins, suggesting that conserved hairpins from the RNAz screen have specific features that make them more different from a miRNA precursor than a random hairpin. In a sense we are not only looking at the structural constraints that allow an RNA molecule to be identified by the RNAi pathway but we are also considering the constraints that allow an RNA molecule to dodge this pathway.
STRUCTURAL FEATURES OF miRNA PRECURSOR STEMS
As miRNAs can be distinguished from other conserved stem–loops by their secondary structure, we were interested in knowing which particular structural features belong solely to miRNAs and how important these elements are in the selection of true positives. We considered commonly used factors such as free energy and the number of consecutive base-pairs as well as more specific information on the position and size of bulges and loops. For each structural parameter, we compared values in the miRNA-enriched cluster and in other hairpin structures dispersed in the tree, and statistical differences were measured using a two-way t-test. The results are shown in Table 1. Surprisingly, free energy and the number of base pairs, the structural criteria often used to fetch out miRNA precursors (Grad et al. 2003; Lai et al. 2003; Adai et al. 2005; Chatterjee and Chaudhuri 2006), are not the most informative when it comes to distinguishing precursors from other conserved hairpins: The P values for these parameters are 2e-3 and 0.21, respectively. The most discriminating features are clearly related to the presence of internal and bulge loops. The number and size of the bulges set the precursors and nonprecursors apart with a P value of 2.2e-16. The number and size of the internal loops are also important (1.6e-7 and 3.8e-10, respectively), as well as the size of the apical loop (6.8e-10). Although true precursors do not differ significantly from other conserved hairpins in terms of overall size and free energy, they tend to display fewer bulges and internal loops, and when such loops or bulges occur, their size is strongly reduced. This suggests that true miRNA precursors tend to minimize the structural features that would confer a more irregular or asymmetric shape to the overall stem–loop.
SECONDARY STRUCTURE CLUSTERING AS A miRNA PREDICTION TOOL
We used the above scoring metric, which compares a candidate structure to both real precursors and other conserved hairpins, as a miRNA prediction method. Using an arbitrary clustering score threshold of >90 for positive hits, we tested 322 human and 79 Caenorhabditis briggsae miRNA precursors from the miRNA registry that were not in the initial tree. We compared this structure comparison method to other, currently used algorithms such as BLAST (Altschul et al. 1990), miRAlign (Wang et al. 2005), and the recently published triplet-SVM classifier (Xue et al. 2005). Table 2 summarizes the performance measures of these algorithms applied to the above data sets of known precursors. One thousand random stem–loops (<−20 Kcal mol−1/15 bp) were extracted from either the human or the C. briggsae genome to compute false-positive rates for each algorithm. BLAST, miRAlign, and the triplet-SVM classifier were given training data sets containing all animal miRNA precursors from miRbase, excluding specific subsets chosen to test prediction accuracy in different contexts. Two different training sets were used to simulate high or low similarity search conditions. For human precursor prediction, training set 1 was deprived of human sequences and training set 2 was deprived of all primate sequences. Likewise, for tests on C. briggsae precursors, training set 1 was deprived of C. briggsae precursors and training set 2 was deprived of both C. briggsae and Caenorhabditis elegans precursors. BLAST does not use a training set per se; instead, each sequence in the set was used in turn to perform a BLAST search against the positive or negative database.
Secondary structure clustering correctly identified 256 of the 322 human precursors (sensitivity = 0.8) and 63 of the 79 C. briggsae precursors (sensitivity = 0.79) with false-positive rates of 0.24 and 0.25, respectively. These values compare well with the other methods, especially when more distantly related training sets (set 2) are used, in which case one in 10 miRNAs is found by miRAlign and even less by BLAST. These results show that the secondary structure alone can determine miRNA precursor identity as strongly as the structure + sequence-based characteristics used in miRNA prediction algorithms. In addition, characteristics learned from human RNA structures can be applied directly to the classification of miRNAs from a distant species such as C. briggsae in the absence of further training.
CERTAIN PRECURSOR FEATURES MAY BE UNDERREPRESENTED IN OTHER ncRNAS
Scanning genomic DNA for potential stem–loops produces in the order of one 33-bp duplex every 10 kb, or 300,000 candidates in the human genome. Further screening is necessary to converge on a tractable set of miRNA candidates. The most successful screens have involved expression data (Bentwitch et al. 2005) or sequence conservation at and around miRNA loci (Grad et al. 2003; Lai et al. 2003). In contrast, knowledge of miRNA precursor structure has remained of little help, as few primary or secondary constraints on pri- or pre-mRNA have been established. The present analysis shows that secondary structure alone is an important aspect of miRNA precursor identity and could be incorporated into miRNA screening procedures. We observed that the most important secondary structure determinants of miRNA precursors are a lower number and reduced size of bulges and internal loops. This suggests that long stem–loops that are asymmetrical or contain several hinges are not as good RNAse III substrates as regular and symmetrical helices. Furthermore, genomic stem–loops that are predicted to be expressed as ncRNAs are more distant in secondary structure from miRNA precursors than randomly selected genomic stem–loops. This observation is interesting in that it suggests a tendency for functional ncRNA to escape the RNAi pathway, possibly through selection of less regular and symmetrical helical structures. Secondary structural features, combined with other features such as tertiary folds or the formation of multiple subunit assemblies, may thus contribute to protect functional ncRNA from unwanted RNAse III cleavage in the nucleus and cytoplasm.
SUPPLEMENTAL DATA AND SOFTWARE AVAILABILITY
A Web site has been set up where users can submit precursor candidates to the scoring procedure described in this article, available at http://tagc.univ-mrs.fr/mirna/. Supplemental material is available at http://tagc.univ-mrs.fr/pub/.
FIGURE 3.
Distributions of miRNA classification scores. Blue indicates scores of 322 known RFAM miRNAs (negative control: diffuse hairpins). Red indicates scores of a set of 1000 randomly selected nonconserved hairpins (ΔG <−20 kcal/mol) from the human genome (negative control: diffuse hairpins). Purple indicates scores of 322 known RFAM miRNAs (negative control: 1000 hairpin structures above).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.366507.
REFERENCES
- Adai, A., Johnson, C., Mlotshwa, S., Archer-Evans, S., Manocha, V., Vance, V., Sundaresan, V. Computational prediction of miRNAs in Arabidopsis thaliana . Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Chatterjee, R., Chaudhuri, K. An approach for the identification of microRNA with an application to Anopheles gambiae . Acta Biochim. Pol. 2006;53:303–309. [PubMed] [Google Scholar]
- Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., Long, J., Stern, D., Tammana, H., Helt, G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Cui, C., Griffiths, A., Li, G., Silva, L.M., Kramer, M.F., Gaasterland, T., Wang, X.J., Coen, D.M. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad, Y., Aach, J., Hayes, G., Reinhart, B., Church, G., Ruvkun, G., Kim, J. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S. miRBase: The microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Han, J., Lee, Y., Yeom, K.H., Nam, J.W., Heo, I., Rhee, J.K., Sohn, S.Y., Cho, Y., Zhang, B.T., Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Höchsmann, M., Toller, T., Giegerich, R., Kurtz, S. Local similarity in RNA secondary structures. Proc. IEEE Comput. Soc. Bioinform. Conf. 2003;2:159–168. [PubMed] [Google Scholar]
- Höchsmann, M., Voss, B., Giegerich, R. Pure multiple RNA secondary structure alignments: A progressive profile approach. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2004;1:53–62. doi: 10.1109/TCBB.2004.11. [DOI] [PubMed] [Google Scholar]
- Hofacker, I., Fekete, M., Stadler, P. Secondary structure prediction for aligned RNA sequences: Technical report. Santa Fe Institute; Santa Fe, NM: 2001. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., Tomancak, P., Williams, R.W., Rubin, G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I.H., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., Bartel, D.P. The microRNAs of Caenorhabditis elegans . Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae, I.J., Zhou, K., Li, F., Repic, A., Brooks, A.N., Cande, W.Z., Adams, P.D., Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., Sewer, A., Lagos-Quintana, M., Sheridan, R., Sander, C., Grasser, F.A., van Dyk, L.F., Ho, C.K., Shuman, S., Chien, M., et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Steffen, P., Voss, B., Rehmsmeier, M., Reeder, J., Giegerich, R. RNAshapes: An integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.H., Gaasterland, T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Zhang, J., Li, F., Gu, J., He, T., Zhang, X., Li, Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- Washietl, S., Hofacker, I.L., Lukasser, M., Huttenhofer, A., Stadler, P.F. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Xue, C., Li, F., He, T., Liu, G.P., Li, Y., Zhang, X. Classification of real and pseudo-microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y., Cullen, B.R. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]