Abstract
Francisella tularensis subverts the immune system to rapidly grow within mammalian hosts, often causing tularemia, a fatal disease. This pathogen targets the cytosol of macrophages where it replicates by using the genes encoded in the Francisella pathogenicity island. However, the bacteria are recognized in the cytosol by the host's ASC/caspase-1 pathway, which is essential for host defense, and leads to macrophage cell death and proinflammatory cytokine production. We used a microarray-based negative selection screen to identify Francisella genes that contribute to growth and/or survival in mice. The screen identified many known virulence factors including all of the Francisella pathogenicity island genes, LPS O-antigen synthetic genes, and capsule synthetic genes. We also identified 44 previously unidentified genes that were required for Francisella virulence in vivo, indicating that this pathogen may use uncharacterized mechanisms to cause disease. Among these, we discovered a class of Francisella virulence genes that are essential for growth and survival in vivo but do not play a role in intracellular replication within macrophages. Instead, these genes modulate the host ASC/caspase-1 pathway, a previously unidentified mechanism of Francisella pathogenesis. This finding indicates that the elucidation of the molecular mechanisms used by other uncharacterized genes identified in our screen will increase our understanding of the ways in which bacterial pathogens subvert the immune system.
Keywords: macrophage, pathogenesis, ASC, caspase-1
Francisella tularensis is a highly infectious, Gram-negative bacterial pathogen that causes the disease tularemia (1). Human infection with F. tularensis usually occurs through cutaneous inoculation after an insect bite or an abrasion. The bacteria evade the host immune system and grow in the cytosol of macrophages. They spread from the site of infection to the regional lymph nodes, causing lymphadenopathy, and disseminate systemically to the spleen, liver, and lungs, where they rapidly grow and overwhelm the host's immune defenses.
F. tularensis subsp. tularensis is the most virulent subspecies of Francisella and is found throughout North America and in parts of Europe. F. tularensis subsp. novicida can cause severe disease in immunocompromised humans but rarely does so in immunocompetent individuals. However, F. novicida shares the same families of virulence genes as F. tularensis, causes a similar disease in mice (2), and thereby serves as a good experimental model with which to study Francisella pathogenesis.
Francisella inhibits the host's protective NF-κB response and proinflammatory signaling after infecting unactivated macrophages (3). Phagocytosed bacteria escape the phagosome before phagosome/lysosome fusion and subsequently replicate in the cytosol (4–7). In activated macrophages, however, the ability of Francisella to escape the phagosome and replicate in the cytosol correlates with the activation of the inflammasome, which contains the host proteins ASC and caspase-1. Inflammasome activation is critical for host defense and leads to the induction of cell death in infected cells and the concomitant release of the proinflammatory cytokines IL-1β and IL-18 (8, 9). These responses may function to limit Francisella's intracellular growth niche and activate the powerful host innate immune system.
Some of the genes Francisella uses to cause disease are known, including the genes in the Francisella pathogenicity island (FPI), which are essential for intracellular replication and the induction of cell death in macrophages as well as for bacterial growth in mice (10–15). The transcription factor MglA regulates all of the genes in the FPI and is also required for replication in macrophages and mice (10, 15, 16). However, the genes required for many other aspects of Francisella pathogenesis (spread from peripheral to systemic sites, phagosome escape within host cells) remain unknown.
Genetic screens are often very effective at identifying virulence factors of bacterial pathogens in vivo. Signature-tagged mutagenesis (STM) identified the Salmonella pathogenicity island SPI2, which is essential for virulence (17). A recently developed microarray-based negative-selection technique called transposon site hybridization (TraSH) or microarray tracking of transposon mutants (MATT) was used to identify virulence factors in Mycobacterium, Salmonella, and Helicobacter (18–20).
Here, we use a microarray-based negative selection technique to identify 164 genes required for F. novicida growth and survival in a mouse model of infection. The screen identified almost all of the known Francisella virulence genes including FPI genes, LPS O-antigen, and capsule biosynthetic genes. We identified genes including a previously uncharacterized class of Francisella virulence factors that we show suppress a host innate immune defense pathway. The genes identified in our screen will serve as a foundation with which to unravel virulence strategies of bacterial pathogens.
Results
Construction of a F. novicida Transposon Insertion Library.
We constructed a transposon insertion library of 12,600 F. novicida mutants, which represents ≈7× coverage of the genome. We used a Tn5-based transposon (21) containing T7 promoters facing outwards at each end of the transposon to facilitate the use of the library in a microarray-based negative-selection screen. The promoter driving the constitutively expressed omp26 gene from F. novicida was used to ensure high expression of the kanamycin resistance cassette within the transposon (21). The presence of only one transposon insertion per clone was verified by Southern blot (data not shown). These results are in agreement with those of Gallagher et al. (21).
Francisella Encounters a Bottleneck for Systemic Spread After s.c. Infection.
To identify genes involved in Francisella pathogenesis, we performed an in vivo negative selection screen. Mice were injected s.c. with 2 × 105 cfu of the transposon insertion library to mimic the most common route of infection through the skin. Genomic DNA was collected from the input bacteria as well as the bacteria harvested from the spleens of five mice at 48 h after infection, digested, and used to generate probes for hybridization to our Francisella microarray (15). Similar to previous experiments using wild-type bacteria, an average of 5.2 × 106 cfu (or ≈108cfu/g of spleen) were present per spleen (8). However, we found that only a very limited repertoire of transposon mutants was isolated from the spleen of each mouse [see supporting information (SI) Fig. 5]. Furthermore, this repertoire of clones was different in each mouse, suggesting that the presence of any given clone in the spleen was a random event and not due to a selective pressure. There is, in effect, a “bottleneck” in the passage of Francisella from the skin to systemic sites such as the spleen, which prevents the use of negative selection techniques following this route of infection.
Negative Selection of Mutants in the Spleen After i.p. Infection.
To circumvent this bottleneck and determine which genes are required for bacterial replication and survival at systemic sites, we infected mice i.p. with the transposon insertion library. Two separate experiments were performed in which the bacteria surviving in the spleens of five mice, 48 h after infection were pooled and compared with bacteria in the input sample taken at the time of infection. Significance analysis of microarrays (SAM), a rigorous statistical test, was used to analyze the microarray data (22). We determined that transposon insertion mutants representing 164 genes (≈9% of the genome) were present in the input pool but absent from the spleens of infected mice in both experiments (SI Table 1). We refer to these genes as negatively selected genes.
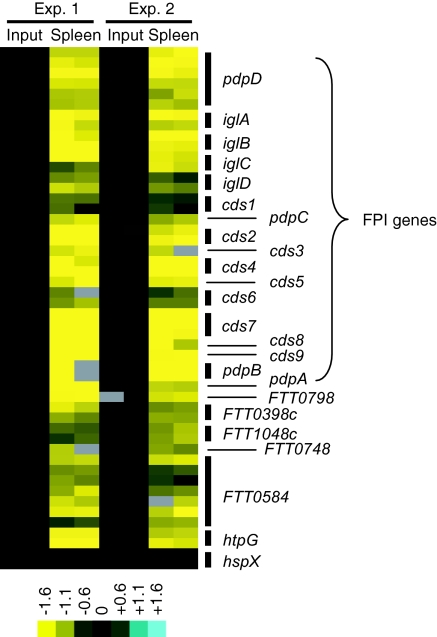
The negative selection method we used here identified all the genes within the FPI, expected metabolic genes, and genes known to be involved in the virulence of other pathogens. We believe these results validate the sensitivity and utility of this genetic screen (Fig. 1 and SI Table 1). Genes involved in metabolism (purine/pyrimidine synthesis, biotin synthesis, and amino acid transport), the stress response (FTT0356/htpG) and genes contributing to the outer surface of the bacteria (capsule and LPS O-antigen synthesis) were identified as being required for Francisella growth and survival in vivo. Genes encoding histidine kinases (FTT0094c/qseC and FTT1736c/kdpD), which may be parts of two-component gene regulatory systems and which were not previously known to play a role in Francisella virulence, were identified. Perhaps of most interest, 44 genes of unknown function or without known orthologs in other microorganisms were negatively selected. These may include classes of virulence genes that participate in previously uncharacterized mechanisms of bacterial pathogenesis.
Fig. 1.
Array data for genes targeted in this study. Bacteria surviving in spleen samples 48 h after infection from two groups of five mice were collected. The transposon mutants present in each sample were identified by microarray analysis. Data for a subset of the negatively selected transposon insertion mutants is shown. Inputs are shown as controls. Blue represents overabundance of signal compared with the input, and yellow signal represents absence relative to the input. Multiple rows for a given gene represent multiple spots on the array.
In Vivo Validation of Negatively Selected Genes.
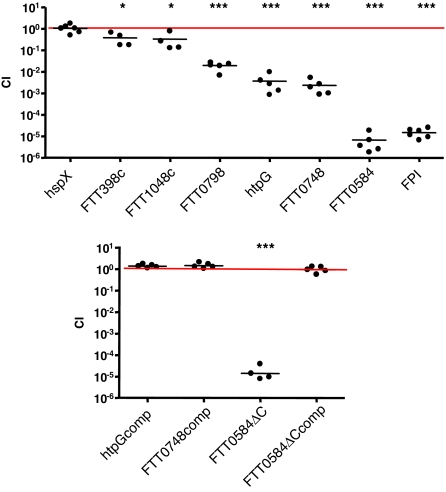
We used SAM to determine which genes were most significantly negatively selected and chose several of these to study, with special emphasis on the genes of unknown function. We made targeted mutants of FTT0398c, FTT0584, and FTT1048c (genes with no characterized orthologs), FTT0748 (homology to the IclR family of transcriptional regulators), mutants in known genes including htpG (stress response), FTT0798 (outer surface/capsule production), and the entire FPI (replication in macrophages). None of the targeted mutants had growth defects in rich broth as compared with the wild-type strain (data not shown) and we verified the deletion of the targeted gene by PCR (SI Fig. 6) and sequencing of the PCR product. We performed competition experiments in which mice were infected s.c. (Fig. 2) or via the i.p. route (SI Fig. 7) with a 1:1 mixture of wild-type strain U112 and one of the mutant strains. Forty-eight hours after infection, the spleens were collected, and the number of wild-type and mutant bacteria in each sample was enumerated.
Fig. 2.
Targeted mutants in negatively selected genes are attenuated in vivo. Groups of four to six mice were s.c. infected with a 1:1 mixture of wild-type bacteria and the indicated bacterial strain. The data represent the CI value for cfu of mutant/wild-type in the spleen 48 h after infection. Bars represent the geometric mean CI value for each group of mice. Each experiment was performed at least twice. ∗, P < 0.05; ∗∗∗, P < 0.0005.
The FPI deletion strain was highly attenuated in mice, similar to mutants in individual FPI genes, which are unable to replicate in macrophages (Fig. 2) (4, 10–13, 15). The FTT0798 (capsule production) mutant was between 10- and 100-fold less virulent compared with wild-type bacteria, a milder phenotype than that of the FPI mutant. This suggests that, similar to other bacterial pathogens and in agreement with the results of Sandstrom et al. (23), the presence of a capsule is important for Francisella virulence.
The htpG (heat shock protein) mutant was strongly attenuated in the spleen (≈1,000-fold) (Fig. 2). In contrast, a mutant in a heat shock protein gene that was not negatively selected, hspX, colonized the spleen of mice similarly to wild-type bacteria. This suggests that specific, but not all, stress response proteins are necessary for Francisella pathogenesis.
The FTT0398c and FTT1048c (unknown function) mutants were mildly attenuated (Fig. 2). In contrast, the FTT0748 (IclR family) mutant was strongly attenuated in the spleen (≈1,000-fold). The FTT0584 (unknown function) mutant was severely attenuated (>5 logs) in the spleen, similar to the phenotype of the FPI mutant (Fig. 2). The severity of the phenotype of the FTT0584 mutant suggests that FTT0584 plays an essential role in Francisella virulence, apparently as important as the known requirement of the FPI for macrophage replication.
We decided to focus on the mutants with the most severe attenuation, htpG, FTT0748, and FTT0584 for further characterization. To ensure that the attenuation of each mutant was due to the deletion of the targeted gene, we complemented the mutants with a wild-type copy of the respective gene (SI Fig. 6). We complemented the mutants in cis, rather than in trans, to ensure that the complementing gene would be retained throughout the course of the in vivo infection in the absence of antibiotic selection. Virulence was restored to both the htpG and FTT0748 mutants upon complementation, because equal numbers of wild-type and complemented bacteria [competitive index (CI) ≈ 1] were present in the spleens of infected mice (Fig. 2). Instead of complementing the full-length FTT0584 (4.9 kb) mutant, we complemented a C-terminal mutant (FTT0584ΔC). The FTT0584ΔC mutant had the same dramatic phenotype as the FTT0584 full-deletion mutant (CI = 10−5; Fig. 2) and regained virulence when the C-terminal region of the gene was replaced (Fig. 2).
Taken together, the in vivo results demonstrate the effectiveness of the genetic screen, because all of the mutants tested were attenuated. To further understand the roles of htpG, FTT0748, and FTT0584 in Francisella virulence, we tested the mutants in in vitro infection experiments in macrophages.
Phenotypes of in Vivo Attenuated Mutants in Macrophage Infections in Vitro.
Macrophages are one of the primary niches for Francisella replication in the host (1). To determine whether defects in intracellular replication were the basis for the observed attenuation of the mutants in mice, we infected bone marrow-derived macrophages with each mutant and assayed for bacterial uptake and replication. All the mutants entered macrophages at similar levels as wild-type bacteria (data not shown). Similar to the results we obtained in mouse infection studies, the FPI and htpG mutants exhibited a strong defect in intracellular growth, whereas the htpG-complemented strain and the hspX mutant replicated with similar kinetics to the wild-type strain (Fig. 3A).
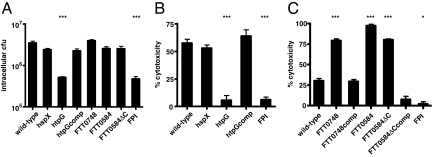
Fig. 3.
Macrophage replication and cytotoxicity assays reveal a class of Francisella virulence factors. Bone marrow-derived macrophages were infected with 10 bacteria per macrophage of the indicated bacterial strains. (A) Cells were lysed and bacteria were collected at 10 h after infection and plated for enumeration. (B and C) Macrophages were activated with heat-killed F. novicida for 12 h before infection. Cell death was quantified by LDH release at 7 (B) or 6 (C) h after infection. Data are representative of three independent experiments. Statistical significance as compared with wild-type: ∗, P < 0.05; ∗∗∗, P < 0.0005.
The FTT0748, FTT0584, and FTT0584ΔC mutants grew to wild-type levels in macrophages (Fig. 3A). Therefore, their dramatic growth/survival defects in the mouse (Fig. 2) do not appear to be due to an inability to replicate within these cells. Taken together, our negative selection screen identified at least two classes of Francisella virulence genes: those in which attenuation in animals and reduced intracellular growth in macrophages parallel one another (htpG and FPI) and those which replicate normally in macrophages but are severely attenuated in the mouse infection model (FTT0748 and FTT0584).
We were surprised to discover that macrophages infected with the FTT0748, FTT0584, and FTT0584ΔC mutants began to die during the course of the replication experiments, a phenomenon not seen in macrophages infected with wild-type bacteria. We performed macrophage cytotoxicity experiments to further investigate this observation.
FTT0748 and FTT0584 Suppress Francisella-Induced Macrophage Cell Death.
Francisella induces cell death in macrophages, which is more rapid in prestimulated versus unstimulated cells (8, 24). To measure cell death in the most robust assay, we infected prestimulated macrophages with wild-type or mutant bacteria and monitored cytotoxicity using the LDH release assay and nuclei staining with the membrane impermeant dye ethidium homodimer-2 (EthD-2). In agreement with our visual observations in unstimulated cells, the FTT0748, FTT0584, and FTT0584ΔC mutants induced LDH release and cell death much earlier than the wild-type strain in prestimulated cells (Fig. 3C and SI Fig. 8). The complemented FTT0748 and FTT0584ΔC mutants induced cell death to similar levels as the wild-type strain. The FPI and htpG mutants did not induce cell death, whereas the htpG complemented and hspX mutant strains induced wild-type levels of cytotoxicity (Fig. 3B).
One explanation for the increased levels of cell death induced by the FTT0748 and FTT0584 mutants could be that they reach the macrophage cytosol faster than wild-type bacteria. Francisella localizes to a LAMP-1+ vacuole before escaping into the cytosol, and colocalization with LAMP-1 can be used to discriminate between vacuolar and cytosolic bacteria (6). Immunofluorescence microscopy revealed that the kinetics of association with LAMP-1 was similar for wild-type bacteria and the FTT0748 and FTT0584 mutants (SI Fig. 9), excluding faster vacuolar escape as an explanation for the rapid cell death induced by these mutants.
Thus far, host cell death has been closely linked with bacterial replication because mutants unable to replicate in macrophages do not induce cell death and mutants which replicate faster than wild-type bacteria induce cell death more rapidly (8, 15). The FTT0748 and FTT0584 mutants are the first Francisella mutants described that are hypercytotoxic even though they replicate to similar levels as wild-type bacteria in unstimulated (Fig. 3A) and prestimulated (data not shown) macrophages. This finding led us to consider the possibility that this pathogen may possess a mechanism to suppress macrophage cell death.
Mechanism of Cell Death Induced by FTT0748 and FTT0584 Mutants.
Macrophage cell death induced by Francisella depends on the host adaptor protein ASC and the cysteine protease caspase-1 (8). To determine whether the rapid cell death induced by the FTT0748 and FTT0584 mutants was due to activation of the ASC/caspase-1 pathway, we infected caspase-1−/− and ASC−/− macrophages with these bacteria. Neither the wild-type nor either of the mutant bacteria induced cell death in caspase-1−/− or ASC−/− macrophages, demonstrating that the cell death induced by the FTT0748 and FTT0584 mutants depends on ASC and caspase-1 (Fig. 4A).
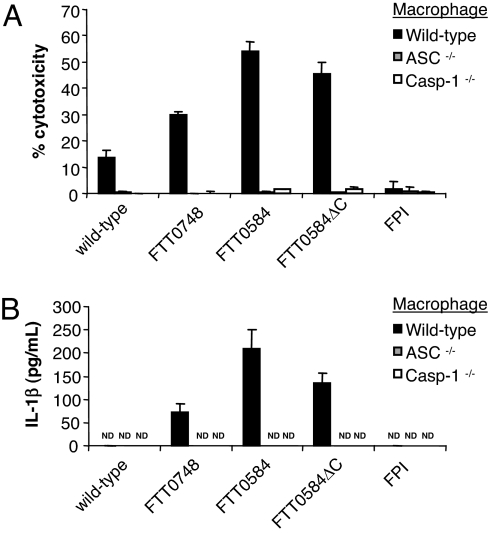
Fig. 4.
Rapid cell death and IL-1β release induced by the FTT0748 and FTT0584 mutants depends on host caspase-1 and ASC. Wild-type, caspase-1−/− and ASC−/− bone marrow-derived macrophages were activated with heat-killed F. novicida for 12 h and then infected with 10 bacteria per macrophage of the indicated strains. (A) Cell death was quantified by LDH release. (B) IL-1β in the supernatant was quantified by ELISA at 6 h after infection. Data are representative of three independent experiments. ND, not detected.
In addition to their role in signaling for cell death, ASC and caspase-1 are required for the processing and secretion of the proinflammatory cytokines IL-1β and IL-18 from macrophages. The FTT0584 mutant induced IL-1β release from macrophages, as did the FTT0748 mutant that had an intermediate phenotype (Fig. 4B). Wild-type bacteria did not induce the release of measurable amounts of IL-1β at this time point. Wild-type bacteria induced IL-1β secretion from thioglycollate-elicited peritoneal macrophages, but the levels induced by the FTT0748 and FTT0584 mutants were markedly higher under these conditions as well (SI Fig. 10). IL-1β release induced by the bacterial mutants depended on ASC and caspase-1, because macrophages deficient in these host proteins did not secrete IL-1β. Thus, in addition to inhibiting macrophage cell death, FTT0748 and FTT0584 limit the secretion of the proinflammatory cytokine IL-1β.
Discussion
Francisella is a highly infectious pathogen with the ability to rapidly grow within mammalian hosts and to subvert the immune system in numerous ways including growing within macrophages and dendritic cells and evading killing by neutrophils (25, 26). To identify genes required for Francisella growth and survival in vivo, we used a microarray-based negative selection screen. We identified a diverse set of genes, including 44 previously unknown genes, that are required for Francisella pathogenesis. Among these, we discovered a class of Francisella virulence genes that modulate host defenses.
We initially attempted to perform our genetic screen after s.c. infection of mice because this route of infection mimics infection through the skin, the route by which Francisella is most often acquired by humans. We found that there was a bottleneck in the passage of Francisella from the skin to the spleen (SI Fig. 5). Similar bottlenecks have been described for enteric pathogens (27–29) after an oral route of inoculation, and it will be interesting to determine the precise nature of this bottleneck in the future. To bypass the bottleneck, we performed the screen after i.p. infection, which eliminates the stochastic seeding of deeper tissues.
We identified 164 genes (≈9% of the genome) as being essential for Francisella growth and survival in vivo (SI Table 1). All the FPI genes, as well as expected metabolic genes (purine/pyrimidine synthesis, biotin synthesis, amino acid transport), were negatively selected, validating the utility of this genetic screen. Genes involved in capsule (capB/C and FTT0798) and LPS O-antigen (wbt locus) biosynthesis were identified, in agreement with previous results (23, 30). The recently characterized Francisella siderophore biosynthetic operon was negatively selected (31), suggesting that, similar to other intracellular pathogens, iron scavenging is important for survival and replication of Francisella in vivo.
Similar to other “global” genetic screening methods, we did not identify all known Francisella virulence genes. For example, mglA and mglB, which are required for expression of FPI genes and Francisella virulence (10, 15, 16) were not identified in our screen. A closer examination of our data suggests that the absence of these and other genes that may have been missed by our screen is likely because of the stringent cutoff used during our analysis or variability at a small number of spots on our microarray. Additionally, the absence of some genes could result from strictly technical reasons such as degradation of a region of DNA during enzymatic digestion, preventing the generation of a probe for hybridization to the microarray. Furthermore, some mutants may be complemented by other “wild-type” clones in vivo, precluding their identification. Finally, the route of infection may determine which sets of Francisella virulence genes are identified. For example, we identified oppB and FTT1209c as being important for virulence when mice are infected s.c. (15), but neither of these genes was identified by our i.p. screen, nor are they important for virulence after i.p. infection (data not shown). This suggests that some genes may be required for growth and survival at peripheral but not systemic sites. Thus, our estimate of the number of genes required for Francisella virulence is likely an underestimation of the actual number of attenuating mutants. Nevertheless, the screen was efficient in identifying many known and, as we show here, previously unknown Francisella virulence genes.
We used SAM to determine which genes were most significantly negatively selected. We focused on genes with unknown functions or without characterized orthologs in other bacterial genomes as well as genes that represented several aspects of Francisella pathogenesis such as the stress response and production of capsule.
Because several capsule genes were negatively selected, we infected mice with a mutant in FTT0798, a homolog of cpsK from Streptococcus agalactiae, which is involved in the production of the capsule (32). FTT0798 was attenuated in vivo (Fig. 2), in agreement with reports on the importance of the capsule in Francisella pathogenesis (23). The htpG mutant, lacking a heat shock protein with homology to the hsp90 chaperone, was attenuated in vivo and was unable to replicate and induce cytotoxicity in macrophages (Figs. 2 and 3), in agreement with a recent study (11). Another heat shock protein gene, hspX, which was not identified in our screen, was not required for Francisella virulence in the assays mentioned, demonstrating that there is specificity as to which stress response genes are involved in infection.
We made deletion mutants for 4 of the 44 hypothetical genes that were identified in our screen, all of which were attenuated after both s.c. and i.p. infection in vivo. Two of the mutants, FTT0398c and FTT1048c, exhibited relatively minor, albeit statistically significant, virulence attenuations (Fig. 2). The FTT0748 mutant was 1,000-fold attenuated as compared with wild-type bacteria. Similar to the FPI mutant, the FTT0584 mutant was severely attenuated (5 logs) in vivo (Fig. 2). The severity of the attenuation of the FTT0584 mutant suggests that FTT0584 is essential for Francisella virulence.
As a first step in determining how FTT0584 contributes to virulence, we performed in vitro macrophage infection experiments. Interestingly, despite its significant attenuation in vivo, the FTT0584 mutant replicated similarly to wild-type bacteria in unactivated (Fig. 3A) and activated (data not shown) macrophages. This is in striking contrast to the FPI mutant, which was unable to replicate in macrophages or in mice. Therefore, despite the fact that both the FPI and FTT0584 mutants are similarly attenuated in vivo, they are likely involved in different processes contributing to Francisella pathogenesis. Similar to the FTT0584 mutant, the FTT0748 mutant was able to replicate in macrophages despite being attenuated in vivo. However, it was unclear how FTT0584 and FTT0748 contribute to Francisella virulence.
Activation of the host ASC/caspase-1 pathway leads to macrophage cell death and release of the proinflammatory cytokines, IL-1β and IL-18. This is an important host defense response because ASC and caspase-1 knockout mice are extremely susceptible to Francisella infection (8). Surprisingly, FTT0584 and FTT0748 mutants induced higher levels of cell death and IL-1β release from activated macrophages compared with wild-type bacteria (Figs. 3 and 4). ASC and caspase-1 are required for cell death and IL-1β release induced by the FTT0584 and FTT0748 mutants (Fig. 4), demonstrating that FTT0584 and FTT0748 directly or indirectly prevent the activation of this host defense pathway. These Francisella virulence genes were previously undescribed as playing a role in limiting macrophage cell death and proinflammatory cytokine release independently of controlling bacterial replication in macrophages and may highlight a virulence strategy of Francisella that acts by modulating ASC/caspase-1 activation. The severity of the in vivo attenuation of the FTT0584 and FTT0748 mutants emphasizes the importance of this class of virulence gene.
FTT0584 has limited homology (17% identity, 27% similarity) to a gene in Wolinella as well as one in Legionella, a pathogen that also triggers the ASC/caspase-1 pathway (33–35). The predicted FTT0584 protein does not contain any conserved domains, and there is some variability in the protein between Francisella subspecies. FTT0584 from F. tularensis Schu4 is shorter (1,124 aa) than the F. novicida U112 protein (1,629 aa), which has a ≈500-aa insertion toward its C terminus. In Francisella holarctica LVS, FTT0584 is truncated and divided into two orfs. FTT0748 encodes for a protein that has homology to the IclR family of transcriptional regulators, which have an N-terminal DNA-binding helix–turn–helix motif and a C-terminal domain that interacts with regulatory proteins. This gene is highly conserved among F. tularensis Schu4, F. holarctica LVS, and F. novicida U112.
We use a genomic-scale negative selection screen to identify a diverse set of genes required for Francisella virulence in vivo. We identify two genes, FTT0584 and FTT0748, which are essential for Francisella pathogenesis and are not required for bacterial replication in macrophages but, rather, suppress the host ASC/caspase-1 pathway, which is critical for innate immune defense. Study of the additional genes identified by our screen may lead to further advances in understanding mechanisms used by bacterial pathogens to cause infection and disease and to subvert or circumvent the host's immune defenses.
Materials and Methods
Bacteria.
Wild-type F. tularensis subsp. novicida strain U112 was described (2). Bacteria were grown overnight with aeration in modified tryptic soy broth (TSB; Difco/BD, Sparks, MD) supplemented with 0.02% l-cysteine and kanamycin (Kan) 30 μg/ml or chloramphenicol (Cat) 3 μg/ml, where appropriate.
Transposon Mutant Library.
We modified a Tn5-based transposon containing a Kan-resistance cassette driven by the omp26 promoter encoded on plasmid pLG62a (21) to include outward facing T7 promoters at each end as detailed in SI Materials and Methods. The transposon was amplified by PCR using primers pLG62a1 and pLG62a2 and complexed with purified transposase as detailed by the manufacturer's instructions (Epicentre, Madison, WI). After 5–7 d at 4°C, the complex was electroporated into F. novicida U112 grown to an OD600 of 1.1. Bacteria were allowed to recover for 4 h before being plated on Mueller–Hinton plates containing 30 μg/ml Kan. Colonies were pooled to create the transposon library.
Mice and Francisella Infections.
Female C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME) between 7 and 10 wk of age were kept under specific pathogen-free conditions in filter-top cages at Stanford University and provided with sterile water and food ad libitum. Experimental studies were in accordance with Institutional Animal Care and Use Committee Guidelines. Mice were inoculated with 2 × 105 cfu s.c. in 50 μl of sterile PBS or i.p. in 500 μl of sterile PBS. Spleens were harvested 2 d after infection, homogenized, and dilutions were plated on supplemented Mueller–Hinton (MH) agar plates containing kanamycin (infections with library) or MH plates with and without the appropriate antibiotic (competition experiments). Plates were grown overnight at 37°C, 5% CO2, and colonies were enumerated. After infections with the library, ≈105 cfu were collected in sterile PBS for isolation of DNA. CI values were calculated by using the formula CI = (mutant cfu in output/wild-type cfu in output)/(mutant cfu in input/wild-type cfu in input).
Microarrays.
Genomic DNA was purified from bacterial pellets by phenol-chloroform extraction. Each DNA sample was divided in two and digested separately with BfaI or RsaI (NEB, Ipswich, MA). The digested DNA was used as the template for in vitro transcription with the AmpliScribe T7-Flash transcription kit (Epicentre) following the manufacturer's protocol, except that 2 μg of digested DNA was used, and the reaction was allowed to proceed for 12–16 h. Purified RNA was used in a RT reaction using SuperScript II(–) (Invitrogen, Carlsbad, CA) and random hexamers as primers. cDNA was labeled with amino-allyl dUTP by using the Klenow (exo-) enzyme. The ssDNA containing amino-allyl dUTP from the mouse output or the library input pools were labeled with Cy3 and Cy5, respectively, before hybridization to our Francisella microarray as described (15). All raw data sets are freely available for download from the GEO database.
Data Analysis.
Normalized data were downloaded from the Stanford Microarray Database according to the median log2 Cy5/Cy3 (logRAT2N). Filters for feature quality, including a Cy3 net median intensity of ≥150 and regression correlation of >0.6, were applied. To compare data from separate i.p. infection experiments, each experimental sample was zero-transformed to the input/input control. Features (spots) missing values for ≥40% of the arrays were removed from the data set. The data sets were analyzed with the SAM program, by using the two-class analysis option to identify features that consistently deviated from the input and samples across all of the arrays with a false discovery rate of 1%.
Targeted Mutagenesis.
Mutant and complemented strains were constructed as described (15) by using the primers listed in SI Table 2. Detailed protocols are available in SI Materials and Methods.
Macrophage Infections.
Bone marrow-derived macrophages were prepared and cultured as described (15). For infections, macrophages were plated and allowed to adhere overnight at 37°C, 5% CO2. The next day, the media was removed, bacteria were added, and plates were spun for 15 min at 850 × g at room temperature to synchronize the infection. Cells were incubated for 30 min, washed three times with warm DMEM before addition of DMEM containing M-CSF. For replication experiments, 3.2 × 105 macrophages were seeded per well in 24-well plates (≈100 heat-killed F. novicida per cell were added for experiments with activated macrophages). Macrophages were lysed at 30 min (to measure entry of bacteria) and 10 h (to measure replication) after infection with 1% saponin (Sigma, St. Louis, MO) in water for 5 min, diluted, and plated on TSA to enumerate cfu. For cytotoxicity experiments, macrophages were seeded in 96-well plates at 5 × 104 cells per well and incubated overnight with heat-killed F. novicida. The next day, they were infected with live bacteria as described above, and supernatant was collected at the indicated times after infection to quantify cell death colorimetrically (CytoTox96 LDH-release kit from Promega, Madison, WI) and for measurement of IL-1β (ELISA from R & D Systems, Minneapolis, MN) according to the manufacturer's directions.
Statistical Analysis.
For CI experiments, log10 values of the CI were compared with zero by using the one-sample Student's t test. Macrophage experiments were analyzed by using the Student's unpaired t test.
Supplementary Material
Acknowledgments
We thank Hanza Mathew, Inna Bilis, and Marina Grunina for technical assistance; Larry Gallagher and Colin Manoil (University of Washington, Seattle) for kindly providing pLG62a containing the Tn5-based transposon; Stephanie Malfatti, Kevin McLoughlin, Patrick Chain, and Emilio Garcia (The Lawrence Livermore National Laboratory, Livermore, CA) for providing F. novicida U112 DNA sequences; Stanley Falkow for helpful discussions; Manuel Amieva, Stanley Falkow, and Elizabeth Joyce for critical reading of the manuscript; and Sara Fisher for administrative assistance. D.S.W. was supported by a postdoctoral fellowship from the Giannini Family Foundation, T.H. was supported by a long-term European Molecular Biology Organization fellowship, and J.J.M. was supported by a U.S. Department of Homeland Security Fellowship. This work was supported by National Institutes of Health–National Institute of Allergy and Infectious Diseases Grants R01 AI063302 and AI-65359 (to D.M.M.).
Abbreviations
- CI
competitive index
- FPI
Francisella pathogenicity island
- SAM
significance analysis of microarrays.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609675104/DC1.
References
- 1.Oyston PC, Sjostedt A, Titball RW. Nat Rev Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 2.Owen CR, Buker EO, Jellison WL, Lackman DB, Bell JF. J Bacteriol. 1964;87:676–683. doi: 10.1128/jb.87.3.676-683.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Telepnev M, Golovliov I, Grundstrom T, Tarnvik A, Sjostedt A. Cell Microbiol. 2003;5:41–51. doi: 10.1046/j.1462-5822.2003.00251.x. [DOI] [PubMed] [Google Scholar]
- 4.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjostedt A. Infect Immun. 2003;71:5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemens DL, Lee BY, Horwitz MA. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Proc Natl Acad Sci USA. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. Cell Microbiol. 2005;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- 8.Mariathasan S, Weiss DS, Dixit VM, Monack DM. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavrilin MA, Bouakl IJ, Knatz NL, Duncan MD, Hall MW, Gunn JS, Wewers MD. Proc Natl Acad Sci USA. 2006;103:141–146. doi: 10.1073/pnas.0504271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. Proc Natl Acad Sci USA. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tempel R, Lai XH, Crosa L, Kozlowicz B, Heffron F. Infect Immun. 2006;74:5095–5105. doi: 10.1128/IAI.00598-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray CG, Cowley SC, Cheung KK, Nano FE. FEMS Microbiol Lett. 2002;215:53–56. doi: 10.1111/j.1574-6968.2002.tb11369.x. [DOI] [PubMed] [Google Scholar]
- 13.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, Elkins KL. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golovliov I, Sjostedt A, Mokrievich A, Pavlov V. FEMS Microbiol Lett. 2003;222:273–280. doi: 10.1016/S0378-1097(03)00313-6. [DOI] [PubMed] [Google Scholar]
- 15.Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM. Infect Immun. 2006;74:6642–6655. doi: 10.1128/IAI.01250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baron GS, Nano FE. Mol Microbiol. 1998;29:247–259. doi: 10.1046/j.1365-2958.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- 17.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 18.Sassetti CM, Rubin EJ. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salama NR, Shepherd B, Falkow S. J Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. Proc Natl Acad Sci USA. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandstrom G, Lofgren S, Tarnvik A. Infect Immun. 1988;56:1194–1202. doi: 10.1128/iai.56.5.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai XH, Golovliov I, Sjostedt A. Microb Pathog. 2004;37:225–230. doi: 10.1016/j.micpath.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 25.McCaffrey RL, Allen LA. J Leukoc Biol. 2006;80:1224–1230. doi: 10.1189/jlb.0406287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosio CM, Dow SW. J Immunol. 2005;175:6792–6801. doi: 10.4049/jimmunol.175.10.6792. [DOI] [PubMed] [Google Scholar]
- 27.Mecsas J, Bilis I, Falkow S. Infect Immun. 2001;69:2779–2787. doi: 10.1128/IAI.67.5.2779-2787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes PD, Bergman MA, Mecsas J, Isberg RR. J Exp Med. 2006;203:1591–1601. doi: 10.1084/jem.20060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meynell GG, Stocker BA. J Gen Microbiol. 1957;16:38–58. doi: 10.1099/00221287-16-1-38. [DOI] [PubMed] [Google Scholar]
- 30.Raynaud C, Meibom KL, Lety MA, Dubail I, Candela T, Frapy E, Charbit A. Infect Immun. 2006 doi: 10.1128/IAI.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan JT, Jeffery EF, Shannon JD, Ramakrishnan G. J Bacteriol. 2006;188:3785–3795. doi: 10.1128/JB.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaffin DO, Mentele LM, Rubens CE. J Bacteriol. 2005;187:4615–4626. doi: 10.1128/JB.187.13.4615-4626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, Kuida K, Mariathasan S, Dixit VM, Flavell RA, et al. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 35.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.