Abstract
We used antisense morpholino oligonucleotides (AMOs) to redirect and restore normal splicing of three prototypic splicing mutations in the ataxia-telangiectasia mutated (ATM) gene. Two of the mutations activated cryptic 5′ or 3′ splice sites within exonic regions; the third mutation activated a downstream 5′ splice site leading to pseudoexon inclusion of a portion of intron 28. AMOs were targeted to aberrant splice sites created by the mutations; this effectively restored normal ATM splicing at the mRNA level and led to the translation of full-length, functional ATM protein for at least 84 h in the three cell lines examined, as demonstrated by immunoblotting, ionizing irradiation-induced autophosphorylation of ATM, and transactivation of ATM substrates. Ionizing irradiation-induced cytotoxicity was markedly abrogated after AMO exposure. The ex vivo data strongly suggest that the disease-causing molecular pathogenesis of such prototypic mutations is not the amino acid change of the protein but the mutated DNA code itself, which alters splicing. Such prototypic splicing mutations may be correctable in vivo by systemic administration of AMOs and may provide an approach to customized, mutation-based treatment for ataxia-telangiectasia and other genetic disorders.
Keywords: ataxia-telangiectasia mutated, mutation-based treatment
This study addresses the restoration of normal splicing in three ataxia-telangiectasia mutated (ATM) splicing mutations by exposure of ataxia-telangiectasia (A-T) cells to antisense morpholino oligonucleotides (AMOs). Antisense oligonucleotides have been used to restore pre-mRNA splicing in other disease models (1–8); however, the therapeutic rationale for each varies considerably and, to our knowledge, none has addressed an intranuclear or DNA repair disorder. Furthermore, we believe that A-T offers several advantages for exploring the therapeutic potential of AMOs, such as (i) the availability of many surrogate markers for evaluating ATM function, ex vivo and in vivo, and (ii) a well characterized spectrum of ATM mutations supported by an extensive cell repository of lymphoblastoid cell lines (LCLs) derived from patients with those mutations (9–12). The LCLs allow pre-mRNA mechanisms to be dissected in great detail. The principles gleaned can then be extended to other tissue targets, such as neuronal cells.
A-T is a progressive autosomal recessive neurodegenerative disorder resulting from mutations in ATM (13). This gene includes 66 exons and encodes a 13-kb mature transcript with an ORF of 9,168 nt. The ATM protein is a serine/threonine kinase with >30 phosphorylation targets (14, 15). It affects control of cell cycle checkpoints, repair of dsDNA breaks, responses to oxidative stress, and apoptosis and is a potent tumor suppressor (16–19). ATM is constitutively expressed in all tissues, primarily in the nucleus. So far no therapy exists for this disorder.
ATM protein is not detectable in most A-T patients. A subset of patients with notable protein levels (5–20%) has milder phenotypes (20). Patients with only residual ATM kinase activity have later onset and slower progression of symptoms. Obligate ATM heterozygotes have 40–50% of normal ATM protein levels (21); despite this, they live essentially normal lives. Taken together, these findings suggest that therapeutic benefits in A-T patients might be achieved if even modest increases in functional ATM protein levels could be achieved. In previous studies we were able to induce ATM protein levels in LCLs from A-T patients with nonsense mutations by exposure to aminoglycosides that read through premature stop codons (22). Such mutations occur in ≈30% of A-T patients.
Approximately half of the unique ATM mutations are splicing mutations (9). Most A-T patients have two distinct mutations. Thus, the majority of A-T patients carry at least one splicing mutation. Approximately 30–40% of these splicing mutations result from exonic variants that create new cryptic splice sites or interfere with splicing regulatory elements (9, 10).
Herein we selected three types of splicing mutations to represent major nonclassical aberrant splicing models in A-T patients (11): a 5′ exonic cryptic splice site variant, a 3′ exonic cryptic splice site variant, and a pseudoexon inclusion variant (12). We designed 25-mer AMOs to target and mask the stronger cryptic splice sites, as evaluated by MaxENT estimates (11). We anticipated that this might redirect the use of nearby normal splice sites. We measured the effects of AMOs at both the mRNA and protein levels. ATM kinase activity and radiosensitivity of A-T cells were also evaluated. We show that AMOs can restore normal ATM splicing processes and, despite the very large size of the ATM protein, can induce significant amounts of full-length functional ATM kinase. This normalized the radiosensitive cellular phenotype of A-T cells.
Results
Design of AMOs.
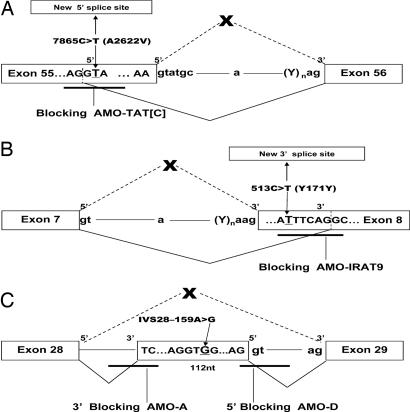
Cell line TAT[C] carried a homozygous mutation, 7865C→T (A2622V). This mutation has been identified in ≈5% of A-T patients in Turkey (11). The mutation creates a new 5′ splice site within exon 55, just upstream of the mutation, which results in a deletion of the last 64 nt (Fig. 1A). We designed the TAT[C]-AMO (TAAGCATCACAAAGTACCTCAACAC) to mask the cryptic 5′ splice site so that the downstream wild-type splice site would be used instead. Cell line IRAT9 contained the homozygous mutation 513C→T(Y171Y), which activates a stronger cryptic 3′ splice site within exon 8 just downstream of the mutation (Fig. 1B) (11). This results in a deletion of the first 22 nt of exon 8. We designed the IRAT9-AMO (AGCCTGAAATACACAGAGAACAATT) to block the aberrant 3′ splice site. Cell line AT203LA contained the mutation IVS28-159 A→G in intron 28 (12). This complex mutation activates both a cryptic 5′ splice site 81 nt downstream of the mutation and a cryptic 3′ splice site 32 nt upstream of the mutation (Fig. 1C), which together result in the insertion of a 112-nt segment of intron 28 in the mRNA. Because we could not anticipate which of the two splice sites of the pseudoexon might have the strongest effect on splicing, we tested two AMO sequences: AT203LA-D (GACAGCTCAAAAGGCATATATTACC) and AT203LA-A (CTGTAGACTGAGAGGAAAAAGTATG), which were designed to mask the 5′ or the 3′ cryptic splice sites, respectively.
Fig. 1.
Schematic representations of three splicing mutations and locations of blocking AMOs. (A) TAT[C] mutation, 7865C→T, results in the deletion of the last 64 nt of exon 55, most likely because of the increased splicing efficiency of AG/GTACTT over AA/GTATGC (wild-type splice site). AMO-TAT[C] was designed to block the mutation-activated cryptic 5′ splice site in exon 55. (B) IRAT9 mutation, 513C→T, results in the deletion of the first 22 nt of exon 8, most likely because of the increased splicing efficiency of ATTTCAG over ACTTCAG or TTTTAAG/AA (wild-type splice site). AMO was designed to block the mutation-activated 3′ splice site in exon 8. (C) Mutation IVS28-159A→G in AT203LA cells results in the inclusion of a portion of intron 28 (112 nt) in the mRNA by activating a downstream cryptic 5′ splice site and/or an upstream cryptic 3′ splice site in intron 28. AT203LA-D and AT203LA-A AMOs were designed to block each of these splicing sites, respectively, to restore normal splicing between exons 28 and 29.
Correction of ATM Pre-mRNA Splicing in TAT[C], IRAT9, and AT203LA Cells by AMOs.
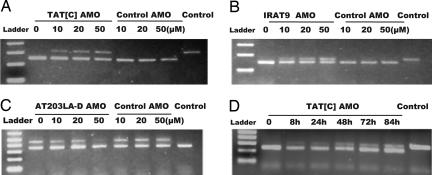
For TAT[C] cells, correction of splicing at the transcriptional level could be observed at 10 μM AMOs, as demonstrated by RT-PCR, with reincorporation of the 64 nt from exon 55. Approximately 40% of pre-mRNA was redirected toward normal/wild-type splicing at 50 μM (Fig. 2A). The normal 5′ splice site was being used after AMO treatment (corrected product, 309 bp; mutant product, 245 bp). Similar effects were observed in IRAT9 cells (Fig. 2B); the normal 3′ splice site was being used after AMO treatment, resulting in the reincorporation of the 22 nt into the transcript (corrected product, 170 bp; mutant product, 148 bp). For AT203LA, whereas the 3′ splice site AMO blocker (AT203LA-A) had no effect (data not shown), the 5′ splice site AMO blocker (AT203LA-D) restored normal splicing (corrected product 344 bp; mutant product, 456 bp). As can be seen in Fig. 2C, at 50 μM concentration of the AT203LA-D AMO, the mutant transcript containing the 112-nt insert (upper band) was almost eliminated, and only the restored normal 344-bp product was observed (lower band). All of the AMO-corrected transcripts in the three cell lines were confirmed by sequencing the PCR products.
Fig. 2.
RT-PCR to demonstrate restoration of ATM mRNA splicing in A-T cell lines after AMO treatment. For A–C, cells were treated with different concentrations of the cognate AMO and a control AMO for 24 h, RNA was isolated, and RT-PCR was performed. Concentrations of AMOs are indicated above the lanes. RNA from normal cells was used as the control for correct splicing (last lanes). As a specificity control, IRAT9-AMO was used as the control for TAT[C] and AT203LA cells; TAT[C]-AMO was used as the control for IRAT9 cells. (A) TAT[C]-AMO was used to treat TAT[C] cells. For RT-PCR, the forward primer was placed at the junction of exon 54 and exon 55; the reverse primer one was in exon 57. The PCR product sizes of the wild-type (corrected) and mutant transcripts were 309 bp and 245 bp, respectively. (B) IRAT9-AMO was used to treat IRAT9 cells. Forward and reverse primers were located in exons 7 and 8, respectively. The PCR product sizes of the wild-type (corrected) and mutant transcripts were 170 bp and 148 bp, respectively. (C) AT203LA-D was used to treat AT203LA cells. Forward and reverse primers were placed in exons 28 and 29, respectively. The PCR product sizes of the wild-type and mutant transcripts were 344 bp and 456 bp, respectively. (D) Time–effect response of TAT[C]-AMO on TAT[C] cells. Cells were incubated with 30 μM TAT[C] AMO. RNA samples were collected at 8, 24, 48, 72, and 84 h and analyzed by RT-PCR.
To evaluate the specificity of each AMO, we crisscrossed the three AMOs among the three cell lines in various experiments. No splicing correction was observed in any of the cell lines exposed to optimal concentrations of noncognate AMOs, indicating that correction of splicing was based on sequence-specific binding for each AMO to the predicted target region of each pre-mRNA. Examples for each AMO are shown as “Control AMO” in Fig. 2 A–C. Additional specificity data are shown in Fig. 3E.
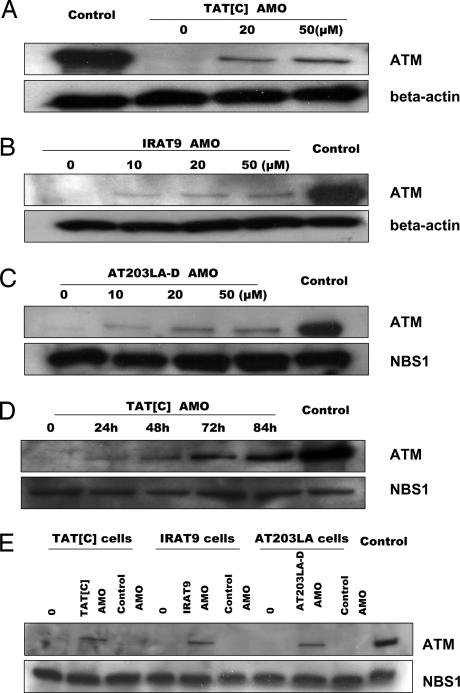
Fig. 3.
Immunoblots of nuclear lysates to demonstrate restoration of ATM protein levels to A-T cell lines after AMO treatment. Cells were treated for 24 h with different concentrations of cognate AMOs. Nuclear protein from normal cells was used as control. NBS1 or β-actin was used as a protein loading control. (A) TAT[C] cells were exposed to TAT[C]-AMO. (B) IRAT9 cells were exposed to IRAT9-AMO. (C) AT203LA cells were exposed to AT203LA-D AMO. (D) TAT[C] cells were exposed to 30 μM TAT[C]-AMO and harvested at different time points. (E) Specificity controls of AMOs. To evaluate the specificity of each AMO on protein expression, the AMOs were crisscrossed among the three cell lines. IRAT9-AMO was used as a control for TAT[C] and AT203LA cells; TAT[C]-AMO was used as a control for IRAT9 cells. For each cell line we treated cells with both the cognate AMO and a noncognate control AMO. No ATM protein was induced in cells exposed to optimized concentrations of noncognate AMOs (50 μM).
We used real-time RT-PCR to further quantify the suppression of mutant transcripts in the cell lines tested [supporting information (SI) Fig. 7A]. After cells were incubated with 50 μM cognate AMOs for 24 h, reduction of mutant mRNA was ≈36% for TAT[C], ≈24% for IRAT9, and ≈56% for AT203LA. This was consistent with the gel-based changes in transcription observed in Fig. 2 A–C.
To determine the time course of AMO effectiveness on ATM transcription, TAT[C] cells were treated with 30 μM AMO, harvested at the times indicated, and assayed by RT-PCR (Fig. 2D). These experiments established that an AMO could redirect aberrant splicing as early as 8 h after treatment (corrected product, 309 bp; mutant product, 245 bp), and this effect lasted at least 84 h, without additional exposure of the cells. Real-time RT-PCR suggested that the maximum effect occurred at ≈48 h (SI Fig. 7B).
Restoration of ATM Protein Synthesis in AMO-Treated Cells.
To determine whether the correction of aberrant splicing also resulted in the translation of ATM protein, we performed Western blots on nuclear extracts from the treated cells. For all three cell lines, ATM protein was induced after 24-h exposures to the cognate AMOs (Fig. 3 A–C). However, although a dose response was seen in each case, we noted that the amount of ATM protein induced by an AMO was not directly proportional to the amounts of transcript observed by RT-PCR data (see Fig. 2). For example, in TAT[C] and IRAT9 cells, whereas ≈40% pre-mRNA was corrected by 50 μM AMOs after 24 h of treatment, <10% ATM protein was induced at the same time point, as compared with the ATM protein level in controls. We considered that, because of the large size of ATM protein, the full effects of an AMO on protein translation might occur hours after maximum transcription levels. To test this, we evaluated ATM protein levels at different time points after exposure to an AMO. Cells were exposed to 30 μM cognate AMOs and collected at different time points. As shown in Fig. 3D, the ATM protein level of TAT[C] cells continued to increase until 84 h. Similar results were observed with IRAT9 and AT203LA cells (SI Fig. 8).
The specificity of cognate AMOs was further addressed with regard to intranuclear ATM protein expression. As can be seen in Fig. 3E, only cognate AMOs induced ATM protein in each of the cell lines.
ATM Kinase Activity Induced by AMO Exposure.
To evaluate ATM kinase activity of the AMO-induced ATM proteins, we tested IR-induced (5 Gy) ATM autophosphorylation at Ser-1981 (ATM-S1981) and phosphorylation/transactivation of substrates SMC1 Ser-957 and p53-Ser-15. As can be seen in Fig. 4, despite the low protein levels observed at 24 h, phosphorylation of all ATM substrates tested was corrected by exposure of the cells to relevant AMOs. The extent of restoration of transactivation for downstream substrates varied; no doubt, these effects would be less variable after longer AMO exposure times.
Fig. 4.
Immunoblots of nuclear lysates to demonstrate restoration of ATM kinase activity of A-T cells after AMO treatment (24 h). Cells were irradiated with 5 Gy and harvested after 30 min. For each cell line, phosphorylation of ATM-S1981, p53-Ser-15, and SMC1-S957 was analyzed. NBS1 or β-actin was used as a protein loading control. A normal LCL was used as a control in each experiment (last two lanes). (A) TAT[C] cells exposed to TAT[C]-AMO. (B) IRAT9 cells exposed to IRAT9-AMO. (C) AT203LA cells exposed to AT203LA-D AMO.
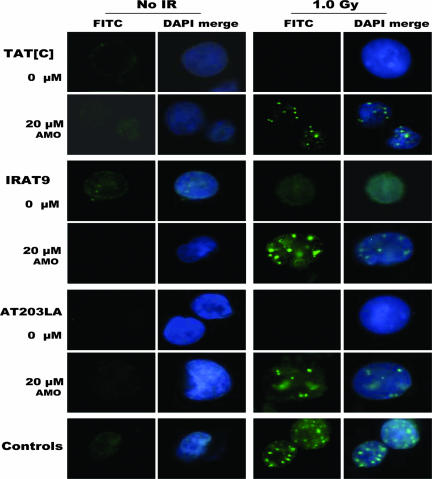
Irradiation-Induced ATM-S1981 Nuclear Foci (IRIF) in AMO-Treated Cells.
As an additional measure of function of AMO-induced ATM proteins, we analyzed immunofluorescence of ATM pS1981 nuclear foci after cells had been exposed to AMOs for 24 h and then damaged by 1-Gy irradiation. This evaluated the presence of ATM protein, its enzymatic ability to autophosphorylate Ser-1981, and the approximate extent of functional correction. After 1 Gy, A-T cell lines exposed to a 20 μM concentration of AMOs showed distinct bright staining of nuclear foci; no foci were observed in unirradiated or untreated cells (Fig. 5). The percentage of IRIF-positive cells was 71% for TAT[C] cells, 62% for IRAT9 cells, and 53% for AT203LA cells. Approximately 85% of wild-type cells (NAT2) showed distinct nuclear foci after irradiation. These pS1981 foci could be almost completely competed out by preincubation of the cells (1 h at 4°C) with the pS1981 epitope (Rockland Immunochemicals, Gilbertsville, PA) against which the antibody was made (data not shown).
Fig. 5.
IR-induced nuclear foci of ATM pS1981 to demonstrate restoration of foci after AMO treatment (24 h). Cells were irradiated with 1 Gy and harvested after 30 min. All three A-T cell lines exposed to cognate AMOs showed significant numbers of nuclear foci as compared with untreated cells. Control cells (wild type) were positive only after IR.
Irradiation-Induced Cell Death Decreased in Cells Exposed to AMOs.
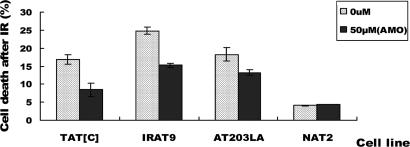
Exposure of A-T cells to ionizing irradiation (IR) results in cytotoxicity. Cell viability was measured by FACS at 48 h after irradiation. The cells had been previously incubated for 24 h with the cognate AMO (50 μM). AMO exposure significantly reduced the percentage of IR-induced cell death in all three cell lines (Fig. 6). The percentages of dead cells for TAT[C] cells decreased from 16.9% to 8.44% after AMO treatment; for IRAT9 cells, cell death was reduced from 24.8% to 15.2%; for AT203LA, cell death was reduced from 18.3% to 13.2% after treatment with AT203LA-D AMO. Exposure of normal cells (NAT2) to the various AMOs showed no toxicity.
Fig. 6.
Cytotoxicity to demonstrate abrogation of radiosensitivity after exposure to cognate AMO. Cells were treated with 50 μM AMO for 24 h and then irradiated with 5-Gy IR. After 48 h cells were harvested and evaluated with propidium iodide staining. A normal LCL (NAT2) was used as a control. The percentage of IR-induced cell death was significantly decreased in all three A-T cell lines after exposure of cognate AMOs.
Discussion
The majority of all inherited diseases result from mutations that cause aberrant splicing; this is proportional to a large extent to the number of exons in a gene (23, 24). In addition, the majority of all human genes undergo alternative splicing to produce splice variants with different functions and specificities. Splice variants have been linked to cancers, as well as to various genetic diseases (25–28). Thus, correction or redirection of pre-mRNA splicing patterns may be potentially applicable to the management of a large variety of diseases.
Antisense oligonucleotides have been used to successfully modulate RNA splicing for cystic fibrosis (CFTR gene), β-thalassemia (β-globin gene), Hutchinson–Gilford progeria syndrome (LMNA gene), and Duchenne muscular dystrophy (Dystrophin gene). These compounds have been used to modify alternative splicing in the SMN2 (spinal muscular atrophy), Bcl-x, and C-myc genes (29–36). Besides blocking cryptic splice sites, antisense oligonucleotides can block splice regulation sequences in the FGFR1 gene (37). They have been delivered systemically to treat mdx mice, a model for Duchenne muscular dystrophy. Direct injection of these antisense agents specifically removed exon 23, which carried a nonsense mutation, and restored a modified dystrophin protein expression level; improved muscle function suggested that exon-skipping of this structural protein was still compatible with function (7). In another study, systemically administered 2′-O-MOE oligonucleotides, formulated in saline, successfully altered splicing of MyD88 in mouse liver (38). On the other hand, the retention of protein integrity and function after exon skipping would not apply for most proteins because they would become unstable and nonfunctional.
In the current study we chose AMOs to correct aberrant disease-causing ATM splicing because of their stability, their high efficiency for binding target RNA, and their long-term activity (at least 84 h in our experiments) (39–41). We have demonstrated splicing correction for three distinct prototypes of splicing mutations. The AMOs successfully blocked mutation-activated cryptic splice sites, either 5′ or 3′ to the mutation and to the wild-type exon/intron junction, and restored correct ATM splicing. The efficiency of ATM correction varied from ≈5% to 25%, depending on the concentration of AMOs and exposure time, and other unexplored factors. We found that the highest ATM protein level was achieved at 84 h after AMO exposure (the longest time point measured), while the maximum correction at mRNA levels was observed around 48 h, indicating a rather long delay between transcription and translation. This would not be unusual for a large protein. It also suggests that turnover of the ATM protein is not particularly rapid and would lend itself well to therapeutic correction.
The restored ATM protein was functional for kinase activity in all three examples, as evaluated by autophosphorylation of ATM, as well as of downstream transactivation the ATM substrates p53 and SMC1. For IRAT9 and AT203LA, the restored ATM protein no longer contained any mutation. However, for TAT[C], the induced ATM protein still contained the missense mutation (A2662V); despite this, it was stable and retained kinase activity, suggesting that the amino acid substitution by itself does not significantly influence ATM function. Moreover, IR-induced cell death of the cells was also reduced after AMO treatment, implying that the induced ATM protein assisted in repair of DNA damage. The safety of inducing such missense proteins will require additional scrutiny in animal models, especially when applied to ATM, because dominant interference has been suggested in some ATM mutagenesis experiments (42) and retroactive induction of apoptosis could target critical cell populations for programmed senescence once ATM levels were restored (17, 18, 43).
The ubiquitous presence of ATM protein in all cells suggests that therapeutic approaches for A-T should target either its increased expression or decreased degradation. To date, only supportive treatment exists, such as physical therapy, speech therapy, and reducing DNA damage with antioxidants, most of which have not been validated by clinical trials. Bone marrow transplantation has been attempted on several occasions but never successfully achieved. Indeed, ablation of an A-T recipient's bone marrow is fraught with difficulties related to the inherent hypersensitivity of A-T cells to IR, radiomimetics, and DNA-damaging agents. Furthermore, a successful bone marrow transplant would not be expected to correct the progressive cerebellar ataxia, ocular apraxia, or dysarthria (slow speech) that represent the major disabilities of these patients. Fetal tissue implantation would be an unlikely avenue for increasing intranuclear ATM levels in existing tissues. Vector-mediated gene therapy models have been described (44); however, unresolved issues include how the ATM gene might be delivered to so many sites and regulated at appropriate times in development.
Our data indicate that AMOs may provide a new approach to customized, mutation-based treatment for A-T patients. These compounds will require both safety and effectiveness testing. However, it is difficult to envision the development of an appropriate animal model for each mutation. It would seem more practical to stratify such testing into protocols that first demonstrate the safety of restoring ATM levels in an ATM-deficient animal model and then test the effectiveness of AMOs on restoration of ATM levels and functions in ATM-deficient animals with a few cognate splicing mutations. Such AMOs would, of course, have to cross the blood–brain barrier in amounts sufficient to restore neural repair. These studies also underscore the importance of properly analyzing and interpreting the molecular mechanisms through which an individual mutation causes disease, because therein lies a logical approach to its correction.
The activity of AMOs appears to be highly dependent on the RNA sequence targeted. For example, in AT203LA cells, the AMO that targeted the 3′ splice site of the pseudoexon (AT203LA-A) had little effect on ATM splicing, whereas the AMO targeting the 5′ splice site (AT203LA-D) showed effective blocking of intron inclusion. The differences in oligonucleotide activity that we observed might also reflect the accessibility of the AMOs to the pre-mRNA sites for hybridization. Once the delivery of AMOs to specific cell types can be achieved, the principles derived from this study can be applied more broadly, and mutation-based customized therapy can be considered for A-T and other genetic disorders.
Materials and Methods
Mutations and Cell Lines.
LCLs TAT[C], IRAT9, and AT203LA were derived from A-T patients with typical phenotypes and carried mutations that were confirmed by bidirectional sequencing of genomic DNA and cDNA (11, 12). They were established by Epstein–Barr virus transformation and were maintained in RPMI medium 1640 with 10% FBS and 1% penicillin/streptomycin at 37°C and 5% CO2. TAT[C] and IRAT9 cells carry homozygous mutations 7865C→T and 513C→T, respectively (11). AT203LA cells have a heterozygous splicing mutation, IVS28-159A→G, which causes the inclusion of two pseudoexons (112 nt and 190 nt; 112 nt is the major isoform); this complex mutation has been previously described (12).
AMO Design and Cell Exposure Conditions.
The 25-mer AMOs were designed to target the aberrant splice site in pre-mRNA for each of the cell lines depicted in Fig. 1. AMOs were synthesized and purified by Gene Tools (Philomath, OR). Endor-Portor (Gene Tools) was used to help cells incorporate AMOs (45). The AMO sequences were as follows (from 5′ to 3′): TAT[C], TAAGCATCACAAAGTACCTCAACAC (mutation site underlined); IRAT9, AGCCTGAAATACACAGAGAACAATT (mutation site underlined); AT203LA-D, GACAGCTCAAAAGGCATATATTACC; and AT203LA-A, CTGTAGACTGAGAGGAAAAAGTATG (neither AMO for AT203LA involves the actual mutation site; see Fig. 1).
For AMO exposure, the cells were resuspended in 5% FBS/RPMI medium 1640 and AMOs were added directly to medium at the concentrations indicated. Endor-Portor (8 μl/ml) was also added to the medium and immediately mixed well. Equal volumes of Endor-Portor were added to untreated cell cultures as controls. Cells were rinsed with PBS and collected at the time points indicated. For addressing the specificity of each AMO, AMOs were used to treat a noncognate cell line.
RNA Preparation, RT-PCR, and Real-Time PCR.
Total RNA was extracted by using the RNeasy kit (Qiagen, Valencia, CA). Reverse transcription reactions were catalyzed by SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). For RT-PCR, specific primers were designed for each mutation to amplify both the full-length and mutant transcripts. The PCR contained 0.2 M of each primer, 1.5 mM MgCl2, 2.0 mM dNTPs, and 1.5 units of Taq and was performed under the following conditions: 36 cycles of 95°C for 30 sec, 52°C for 30 sec, and 72°C for 30 sec after an initial denaturation for 3 min, followed by a final extension for 8 min. Primer sequences were as follows: TAT[C]-FW, TTGATGAGGATCGAACAGAGG; TAT[C]-RE, TTACACCTCCTGCTAAGCGAAATTC; IRAT9-FW, AATGGTGCTATTTACGGA; IRAT9-RE, TAGCCACTAAAACTCTATGAAC; AT203LA-FW, TCTTGTTATAAGGTTTTGATTCC; AT203LA-RE, TCAGTGCTCTGACTGGCACT.
Real-time PCR was performed in a ABI 7700 Sequence Detector using QuantiTect SYBR Green PCR (Qiagen) according to the manufacturer's instructions. ATM mutant transcript-specific (junction) primers were designed for each A-T cell line. GAPDH was used for the relative quantification. Primer sequences are available upon request. The PCR conditions were as follows: 95°C for 15 min, followed by 40 cycles of 94°C for 15 sec, 56°C for 30 sec, 72°C for 30 sec, and finally storage at 4°C.
Western Blots.
For AMO-induced ATM protein analyses, LCLs were treated with various concentrations of AMOs, and nuclear extracts were prepared for each sample, according to NE-PER protocol (Pierce, Rockford, IL). The proteins were separated on a 7.5% SDS polyacrylamide gel, and Western blot analysis was performed by using an anti-ATM antibody (Novus, Littleton, CO) at 1:1,000 dilutions. For ATM kinase activity, cells were first irradiated with 5-Gy gamma IR to induce DNA damage and incubated at 37°C for 30 min before nuclear lysates were prepared. Autophosphorylation of ATM was detected by Western blot using ATM-Ser-1981 antibody (Rockland Immunochemicals); phosphorylation of p53 and SMC1 was detected with anti-p53-Ser-15 (Cell Signaling Technology, Beverly, MA) and anti-SMC1-Ser-957 (Novus) antibodies, respectively, at dilutions of 1:1,000.
Immunofluorescence.
Cells were incubated for 24 h with AMOs, followed by 1-Gy irradiation treatment, and then incubated at 37°C for 30 min. Subsequently, the cells were rinsed with PBS and dropped onto 100 μg/ml poly(d-lysine)-coated coverslips and fixed with 4% paraformaldehyde for 10 min at room temperature, and the nuclei were permeabilized with 0.5% Triton X-100 for 10 min at room temperature. After five rinses in 1× PBS, coverslips were blocked for 1 h at room temperature in 10% FBS, incubated for 1 h at room temperature with mouse anti-ATM pS1981 (Rockland Immunochemicals) at a dilution of 1:500, and rinsed three times in PBS with 0.1% Triton X-100. After a second incubation in 10% FBS for 1 h at room temperature, cells were stained with FITC-conjugated anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) at a dilution of 1:150 for 1 h at room temperature. Coverslips were mounted onto slides by using VECTASHIELD with 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Images were captured with FISH analysis software (Vysis, Downers Grove, IL) connected to a Leica DM RXA automated microscope equipped with Photometrix SenSyn.
Cell Viability Measured by Flow Cytometry.
Cells were treated with AMOs for 24 h and irradiated with 5-Gy IR. After a 48-h incubation, the cells were rinsed with 1× PBS and stained with propidium iodide to assess the number of dead cells. Samples were analyzed by a Becton Dickinson FACScan Analytic Flow Cytometer (Flow Cytometry Core Laboratory, University of California, Los Angeles). Each group of cells analyzed included an unstained control.
Supplementary Material
Acknowledgments
These studies were supported by Public Health Service Grants NS 35322 and AI 067769 and the Ataxia-Telangiectasia Medical Research Foundation.
Abbreviations
- A-T
ataxia-telangiectasia
- LCL
lymphoblastoid cell line
- AMO
antisense morpholino oligonucleotide
- IR
ionizing irradiation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608616104/DC1.
References
- 1.Suwanmanee T, Sierakowska H, Lacerra G, Svasti S, Kirby S, Walsh CE, Fucharoen S, Kole R. Mol Pharmacol. 2002;62:545–553. doi: 10.1124/mol.62.3.545. [DOI] [PubMed] [Google Scholar]
- 2.Friedman KJ, Kole J, Cohn JA, Knowles MR, Silverman LM, Kole R. J Biol Chem. 1999;274:36193–36199. doi: 10.1074/jbc.274.51.36193. [DOI] [PubMed] [Google Scholar]
- 3.Lacerra G, Sierakowska H, Carestia C, Fucharoen S, Summerton J, Weller D, Kole R. Proc Natl Acad Sci USA. 2000;97:9591–9596. doi: 10.1073/pnas.97.17.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scaffidi P, Misteli T. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClorey G, Moulton HM, Iversen PL, Fletcher S, Wilton SD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- 6.McClorey G, Fletcher S, Wilton S. Curr Opin Pharmacol. 2005;5:529–534. doi: 10.1016/j.coph.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J, Jadoon A, Bou-Gharios G, Partridge T. Proc Natl Acad Sci USA. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, den Dunnen JT, van Ommen GJ. Hum Mol Genet. 2001;10:1547–1554. doi: 10.1093/hmg/10.15.1547. [DOI] [PubMed] [Google Scholar]
- 9.Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengut S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA, Concannon P. Am J Hum Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Concannon P, Gatti RA. Hum Mutat. 1997;10:100–107. doi: 10.1002/(SICI)1098-1004(1997)10:2<100::AID-HUMU2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 11.Eng L, Coutinho G, Nahas S, Yeo G, Tanouye R, Babaei M, Dork T, Burge C, Gatti RA. Hum Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho G, Xie J, Du L, Brusco A, Krainer AR, Gatti RA. Hum Mutat. 2005;25:118–124. doi: 10.1002/humu.20170. [DOI] [PubMed] [Google Scholar]
- 13.Perlman S, Becker-Catania S, Gatti RA. Semin Pediatr Neurol. 2003;10:173–182. doi: 10.1016/s1071-9091(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 14.Bakkenist CJ, Kastan MB. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 15.Kurz EU, Lees-Miller SP. DNA Repair (Amsterdam) 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, McKinnon PJ. Apoptosis. 2000;5:523–529. doi: 10.1023/a:1009637512917. [DOI] [PubMed] [Google Scholar]
- 17.Barlow C, Brown KD, Deng CX, Tagle DA, Wynshaw-Boris A. Nat Genet. 1997;17:453–456. doi: 10.1038/ng1297-453. [DOI] [PubMed] [Google Scholar]
- 18.Lavin MF, Spring K. Toxicology. 2002;181–182:483–489. doi: 10.1016/s0300-483x(02)00462-6. [DOI] [PubMed] [Google Scholar]
- 19.Shiloh Y. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Gilad S, Chessa L, Khosravi R, Russell P, Galanty Y, Piane M, Gatti RA, Jorgensen TJ, Shiloh Y, Bar-Shira A. Am J Hum Genet. 1998;62:551–561. doi: 10.1086/301755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chun HH, Sun X, Nahas SA, Teraoka S, Lai CH, Concannon P, Gatti RA. Mol Genet Metab. 2003;80:437–443. doi: 10.1016/j.ymgme.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Lai CH, Chun HH, Nahas SA, Mitui M, Gamo KM, Du L, Gatti RA. Proc Natl Acad Sci USA. 2004;101:15676–15681. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modrek B, Resch A, Grasso C, Lee C. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modrek B, Lee C. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 25.Krawczak M, Reiss J, Cooper DN. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Blanco MA, Baraniak AP, Lasda EL. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 27.Xing Y, Xu Q, Lee C. FEBS Lett. 2003;555:572–578. doi: 10.1016/s0014-5793(03)01354-1. [DOI] [PubMed] [Google Scholar]
- 28.Faustino NA, Cooper TA. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 29.Madocsai C, Lim SR, Geib T, Lam BJ, Hertel KJ. Mol Ther. 2005;12:1013–1022. doi: 10.1016/j.ymthe.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Lim SR, Hertel KJ. J Biol Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 31.Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. Proc Natl Acad Sci USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Nat Biotechnol. 1999;17:1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- 33.Mercatante DR, Bortner CD, Cidlowski JA, Kole R. J Biol Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 34.Mercatante DR, Mohler JL, Kole R. J Biol Chem. 2002;277:49374–49382. doi: 10.1074/jbc.M209236200. [DOI] [PubMed] [Google Scholar]
- 35.Giles RV, Spiller DG, Clark RE, Tidd DM. Antisense Nucleic Acid Drug Dev. 1999;9:213–220. doi: 10.1089/oli.1.1999.9.213. [DOI] [PubMed] [Google Scholar]
- 36.Villemaire J, Dion I, Elela SA, Chabot B. J Biol Chem. 2003;278:50031–50039. doi: 10.1074/jbc.M308897200. [DOI] [PubMed] [Google Scholar]
- 37.Bruno IG, Jin W, Cote GJ. Hum Mol Genet. 2004;13:2409–2420. doi: 10.1093/hmg/ddh272. [DOI] [PubMed] [Google Scholar]
- 38.Vickers TA, Zhang H, Graham MJ, Lemonidis KM, Zhao C, Dean NM. J Immunol. 2006;176:3652–3661. doi: 10.4049/jimmunol.176.6.3652. [DOI] [PubMed] [Google Scholar]
- 39.Summerton J, Weller D. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 40.Summerton J. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 41.Amantana A, Iversen PL. Curr Opin Pharmacol. 2005;5:550–555. doi: 10.1016/j.coph.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Scott SP, Bendix R, Chen P, Clark R, Dork T, Lavin MF. Proc Natl Acad Sci USA. 2002;99:925–930. doi: 10.1073/pnas.012329699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y, Chong MJ, McKinnon PJ. J Neurosci. 2001;21:6687–6693. doi: 10.1523/JNEUROSCI.21-17-06687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cortes ML, Oehmig A, Perry KF, Sanford JD, Breakefield XO. Neuroscience. 2006;141:1247–1256. doi: 10.1016/j.neuroscience.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 45.Summerton JE. Ann NY Acad Sci. 2005;1058:62–75. doi: 10.1196/annals.1359.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.