Abstract
Before the onset of autoimmune destruction, type 1 diabetic patients and an animal model, the nonobese diabetic (NOD) mouse, show morphological and functional abnormalities in target organs, which may act as inciting events for leukocyte infiltration. To better understand these abnormalities, but without the complications associated with lymphocytic infiltrates, we examined genes expressed in autoimmune target tissues of NOD/severe combined immunodeficient (scid) mice and of autoimmune-resistant C57BL/6/scid mice. Our results suggest that the NOD genetic background may predispose them to diabetic complications, including insulin resistance in the absence of high circulating glucose levels and without autoimmune destruction of their β cells. Several of these genes lie within known type 1 and 2 diabetes loci. These data suggest that the NOD mouse may be a good candidate to study an interface between type 1 and type 2 diabetes.
Keywords: insulin resistance, microarray, NSY mice
Type 1 diabetes (T1D) results from the autoimmune destruction of the insulin-producing β cells of the pancreas, which leads to complete insulin deficiency and a concomitant loss of glucose homeostasis. The nonobese diabetic (NOD) mouse approximates human T1D with respect to genetic complexity, the strong contribution to susceptibility of particular MHC class II alleles, and the importance of an autoreactive T cell compartment that, when transferred, is capable of causing diabetes in lymphocyte-deficient NOD/severe combined immunodeficient (scid) mice (NOD mice homozygous for the SCID mutation) (1, 2).
In addition to immunologic defects (3), NOD mice show abnormal pancreatic development, exaggerated β cell death, and altered pancreatic functions before leukocyte infiltration, which precedes the priming of diabetogenic lymphocytes and autoimmune destruction of β cells (reviewed in refs. 4 and 5). Similar to human T1D, multiple tissues including submandibular and lacrimal glands in NOD mice also show abnormalities before autoimmune infiltration (6, 7), and NOD/scid mice show many of the same histological alterations seen in the autoimmune target tissues of NOD mice (5, 6, 8).
It was suggested that these tissue abnormalities result from immune cell deficiencies, especially those of macrophages (reviewed in ref. 4). However, abnormal morphologies and cellular compositions are often indicative of intrinsic alterations in differentiation programs and physiological states of cells. NOD mice were derived from the same colony as Nagoya–Shibata–Yasuda (NSY) mice that spontaneously develop type 2 diabetes (T2D) (9). T2D is caused by functional defects of the β cells and/or insulin resistance. It is therefore possible that the β cell abnormalities in NOD mice are, at least in part, intrinsic.
Here we use microarrays to compare the gene expression patterns of NOD/scid pancreata to those of C57BL/6 (B6)/scid pancreata. To better identify general mediators of tissue abnormalities, which could manifest in shared expression differences among different target tissues, we also analyzed submandibular and lacrimal glands. Mice congenic for the SCID mutation were used to avoid the contributions of T and B cells, both directly from their transcripts and indirectly from their interactions with target tissues during inflammation. Our results suggest that there is common etiology between type 1 and type 2 diabetes and identify genes and pathways that may operate toward the development of autoimmunity and diabetes.
Results and Discussion
In NOD mice, morphological alterations of target tissues are present before the onset of leukocyte infiltration. We therefore analyzed pancreata and submandibular and lacrimal glands from 6-, 9-, and 15-week-old NOD/scid mice, respectively (7, 10, 11). These ages correspond to periods of early inflammation in female NOD mice in the respective tissues. Whole pancreata, instead of just islets, were analyzed because abnormalities were reported outside of the islet proper and in exocrine pancreatic tissues (10).
To reduce the false discovery rate of differentially expressed genes to <5%, five samples were isolated and processed independently for each tissue and each time point. The differential expression of select genes was verified by Northern blots and/or semiquantitative PCR (data not shown).
Although gene expression patterns were similar between the same tissues of NOD/scid and B6/scid mice, cluster analysis of individual submandibular gland samples showed that the five independent NOD/scid expression profiles were distinct from those of B6/scid (data not shown). Among the ≈6,000 to ≈9,000 genes called “present” in these tissues, ≈200 to ≈600 genes exhibited significant and >1.5-fold differences (Fig. 2 and Table 4, which are published as supporting information on the PNAS web site). Approximately one-third of the differentially expressed genes were ESTs. Known/characterized genes were further grouped by function and by whether they showed tissue-specific or tissue-common differences or mapped close to diabetes loci as discussed below.
Altered Pancreatic Endocrine Functions.
As shown in Table 1, there was increased insulin and decreased glucagon expression in NOD/scid pancreata. This finding matched a report that NOD/scid and NOD mice express more preproinsulin and less glucagon than B6 mice (12). Additionally, we found that galanin (a physiological insulin release inhibitor), islet amyloid polypeptide (a secreted molecule that inhibits insulin-stimulated glucose uptake), and the Ras gene family member Q (Rhoq), which plays an important role in insulin-regulated glucose uptake, all showed decreased expression in NOD/scid pancreata. A straightforward prediction is that NOD/scid mice, relative to B6/scid mice, release more insulin and sequester more glucose after glucose challenge and develop insulin resistance.
Table 1.
Partial list of genes that show tissue-specific changes
| Probe set | Gene symbol (name) | Fold change |
|---|---|---|
| Pancreas | ||
| 113815_at | Ctrc* (chymotrypsin C) | 23.2 |
| 102755_at | Itlna* (intelectin a) | 13.7 |
| 97658_f_at | Ins1* (insulin I) | 1.7 |
| 100407_at | Gal* (galanin) | −1.8 |
| 99488_at | Iapp* (islet amyloid polypeptide) | −1.8 |
| 165472_i_at | Gcg* (glucagon) | −1.9 |
| 137506_at | Rhoq (Ras homolog gene family, member Q) | −2.8 |
| 103367_at | Galgt1* (β-1,4-N-acetyl-galactosaminyl transferase 1) | −30.6 |
| Submandibular gland | ||
| 98480_s_at | Ren1, Ren2 (renin 1, renin 2) | 691.0 |
| 101636_at | Spt2 (salivary protein 2) | 128.5 |
| 104496_i_at | Klk8 (kallikrein 8) | 5.7 |
| 98391_at | Klk11 (kallikrein 11) | 3.0 |
| 104497_f_at | Klk8 (kallikrein 8) | 2.5 |
| 168876_f_at | Klk21 (kallikrein 21) | 1.9 |
| 100719_f_at | Klk16 (kallikrein 16) | 1.9 |
| 162305_f_at | Klk1, Klk9 (kallikrein 9, kallikrein 1) | 1.8 |
| 102693_f_at | Klk26 (kallikrein 26) | 1.8 |
| 92606_at | Uox (urate oxidase) | −17.0 |
| Lacrimal gland | ||
| 92837_f_at | Mug1* (murinoglobulin 1) | 5.4 |
| 92601_at | Pnliprp1 (pancreatic lipase-related protein 1) | 3.0 |
| 111006_at | Plcb1 (phospholipase C, β1) | 3.0 |
| 167894_f_at | Lgals8 (lectin, galactose-binding, soluble 8) | 2.2 |
| 96054_f_at | Acp1 (acid phosphatase 1, soluble) | 2.1 |
| 112766_at | Lman1* (lectin, mannose-binding, 1) | 2.1 |
| 99669_at | Lgals1 (lectin, galactose-binding, soluble 1) | −1.7 |
| 166259_f_at | Ctsc* (cathepsin C) | −21.8 |
Genes are grouped according to their tissue origin (pancreas, submandibular gland, and lacrimal gland). Gene symbols, names, probe sets, and fold changes are listed. Fold change is defined as NOD/scid expression divided by B6/scid expression. Fold changes <1 are indicated with a negative sign (−) and are reported as 1/fold change. Loci in humans and mice that are associated with T1D or T2D and map close to the identified genes or their human counterparts are identified by one asterisk for each locus and are shown in Table 3.
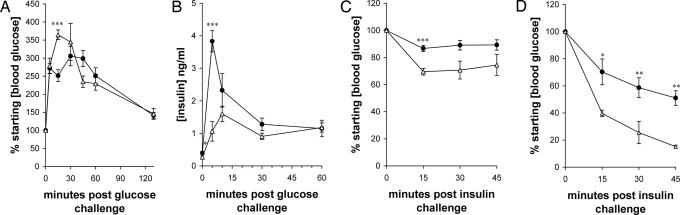
Indeed, 6-week-old NOD/scid mice showed a blunted early rise in blood glucose levels during i.p. glucose tolerance tests but had a similar overall time to blood glucose normalization (Fig. 1A), and NOD/scid mice showed a faster early and larger overall insulin release in such tests (Fig. 1B). As shown in Fig. 1 C and D, the insulin-dependent serum glucose depression in B6/scid mice exceeded that in NOD/scid mice by 15 min and became more pronounced at 30 and 45 min when 2 units/kg insulin was administered. A similar pattern was seen with 0.75 units/kg insulin. Thus, NOD/scid mice are more insulin-resistant than B6/scid mice. Although different inbred strains of mice show different levels of insulin resistance, B6 mice are more insulin-resistant (13) and more prone to T2D than most others. Significantly, the kinetics and the extent of responding glucose levels of NOD/scid mice indicated that NOD/scid mice were more insulin-resistant than mice with a targeted disruption of the insulin receptor substrate 2 gene, which develop T2D (14).
Fig. 1.
NOD/scid and B6/scid mice respond differently in i.p. insulin and glucose tolerance tests. Six-week-old NOD/scid (filled circles) and B6/scid (open triangles) female mice were challenged i.p. with 2 g/kg glucose (A), 3 g/kg glucose (B), 0.75 units/kg insulin (C), and 2 units/kg insulin (D), and their blood glucose concentration (A, C, and D) and serum insulin concentration (B) were monitored over time. Values are means ± SEM. Two-tailed unpaired t tests were then used to analyze the time points, and significant differences are indicated (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
The fact that, compared with B6 mice, NOD mice are insulin-resistant to some degree has been reported before. However, in those experiments 2-month-old mice were analyzed, and at this age the islets of NOD mice are already fully infiltrated by lymplocytes. It was suggested that the production of inflammatory cytokines such as IL-1 in the insulitic islets is a crucial contributing factor to the insulin resistance (15).
NOD/scid mice have normal to low blood glucose levels and higher circulating insulin levels when compared with other inbred strains of mice, yet NOD/scid mice have defects in glucose homeostasis and insulin responses. These results suggest that physiological abnormalities that are commonly associated with diabetes can develop in the absence of high circulating glucose concentrations and without the autoimmune destruction of the β cells. By extension, NOD mice should have similar defects, which could be masked but compounded over time by the actions of autoreactive lymphocytes. In fact, as discussed below, our data suggest that altered gene expression in NOD/scid mice may lead to other complications that are commonly associated with diabetes.
Alteration in Vascular Distensibility.
Vascular abnormalities are prominently associated with all forms of diabetes. In our analysis the most differentially expressed gene was renin (almost 700-fold), which cleaves angiotensinogen to produce the vasoconstrictor angiotensin. This finding indicates that NOD mice are a high-renin strain. In such strains the amount of renin that is synthesized in the submandibular gland far exceeds the amount that is synthesized in the kidney, and there is a direct correlation between protein and mRNA levels. Furthermore, the >100-fold differences in renin levels between high- and low-renin (such as B6) strains are observed after 4 weeks of age and are much more pronounced in female mice (16–18).
Kallikreins are converted in the blood into bradykinins, which are powerful promoters of capillary permeability and leakage. Kallikreins were among the most abundant messages in the submandibular glands of both strains, and most were expressed at even higher levels in the NOD/scid samples. Increased renin and kallikrein expression is in line with the reported abnormal blood flow and increased vascular permeability in NOD target tissues (6). Consistent with this finding, transcripts for hemoglobin were higher in NOD/scid tissues and likely reflected the presence of a greater number of erythrocyte-derived transcripts within them. It is also possible that the hemoglobin loci are linked to diabetes because, whereas human IDDM2 is believed to be insulin, the hemoglobin β-chain complex also maps to that region. In addition, this complex also maps to Nidd1k (19).
Altered Cell–Cell and Cell–Extracellular Matrix (ECM) Interactions.
Altered cell–cell and cell–ECM interactions and endoplasmic reticulum (ER) stress may contribute to altered morphogenesis and increased cell death in NOD pancreata and other autoimmune target tissues. As shown in Table 2, there was much lower expression of thymic shared antigen 1 (TSA-1/Ly6E) in all NOD/scid tissues. TSA-1 knockout mice are embryonic lethal with an abnormal embryonic morphology (20) that is consistent with altered cell–cell and cell–ECM interactions. In addition, Fxyd3 (formerly Mat-8) also showed reduced expression. Fxyd3 has been implicated in ion channel regulation, where it may regulate the Na+ and K+ balance in and around cells (21). There were also large differences in the expression of glycosylation enzymes, suggesting qualitative and quantitative differences in the carbohydrate moieties on glycoproteins and glycolipids (Tables 1 and 2).
Table 2.
Genes that show tissue-common changes
| Probe set | Gene symbol (name) | Fold change |
||
|---|---|---|---|---|
| Pa | SmG | LaG | ||
| Cell–cell/ECM interaction and commmunication | ||||
| 101487_f_at | Ly6e (lymphocyte antigen 6 complex, locus E) | −21.6 | −9.9 | −134.6 |
| 109565_at | Mpp5 (membrane protein, palmitoylated 5) | −1.7 | −1.7 | |
| 103059_at | Fxyd3 (FXYD domain-containing ion transport regulator 3) | −3.0 | −7.4 | |
| 134759_at | St3gal4* (ST3 β-galactoside α-2,3-sialyltransferase 4) | −2.2 | −52.6 | |
| 92655_at | Gcnt1 [glucosaminyl (N-acetyl) transferase 1, core 2] | 1.9 | 2.4 | |
| 98793_at | Gdf5 (growth differentiation factor 5) | 2.9 | 2.7 | |
| 95571_at | Slc30a4* (solute carrier family 30, member 4) | 1.9 | 1.5 | |
| 103918_at | Slc15a2 (solute carrier family 15, member 2) | −2.9 | −2.4 | |
| 104380_at | Slc35a1* (solute carrier family 35, member 1) | 2.1 | 1.6 | |
| 104451_at | Slc11a2* (solute carrier family 11, member 2) | −2.0 | −2.0 | |
| 96353_at | Tmem14c (transmembrane protein 14C) | −4.0 | −5.1 | |
| 114922_at | Garnl4 (GTPase-activated RANGAP domain-like 4) | 2.4 | 1.9 | |
| Protein trafficking and processing | ||||
| 115795_at | Rpgrip1*** (retinitis pigmentosa GTPase regulator interaction protein 1) | −14.8 | −16.3 | −11.0 |
| 103471_at | Tbc1d15 (TBC1 domain family, member 15) | −4.2 | −3.5 | −2.5 |
| 99643_f_at | Cpe (carboxypeptidase E) | −2.0 | −2.4 | −2.5 |
| 96951_at | Atp6v1d (ATPase, H+ transporting, V1 subunit D) | −1.9 | −2.2 | −2.0 |
| 101741_at | Psmb5-ps (proteasome subdomain, β-type 5, pseudogene) | −3.9 | −2.7 | −5.2 |
| 101963_at | Ctsl** (cathepsin L) | −3.9 | −1.6 | |
| 98543_at | Ctss* (cathepsin S) | −2.2 | −3.5 | |
| 110424_at | Ube2b* (ubiquitin-conjugated enzyme E2B) | −1.8 | −1.9 | |
| 93491_f_at | Ube2d3 (ubiquitin-conjugated enzyme E2D 3) | −1.7 | −2.9 | |
| 93281_at | Rcn2* (reticulocalbin 2) | −5.0 | −2.8 | |
| 137557_at | Pigv (phosphatidylinositol glycan, class V) | −4.5 | −3.1 | |
| 93045_at | Abcd3 (ATP-binding cassette, subfamily D, member 3) | −1.9 | −2.3 | |
| 160139_at | Hspb8 (heat shock 27-kDa protein 8) | −2.4 | −2.1 | |
| 98956_at | Tram1 (translocating chain-associate member protein 1) | 1.8 | 2.0 | |
| 100496_at | Pam* (peptidylglycine α-amidating monooxygenase) | 15.6 | 12.6 | |
| 94269_at | Rabac1 (Rab acceptor 1) | −1.6 | −1.6 | |
| 112948_at | Rab6b (RAB6B, member RAS oncogene family) | −2.2 | −3.1 | |
| 104257_g_at | Pscdbp (pleckstrin homol., Sec7 and coiled-coil domain-binding protein) | −1.7 | −1.9 | |
| RNA and chromatin modification | ||||
| 112704_at | Apobec3 (apolipoprotein B-editing complex 3) | −2.9 | −20.0 | −5.5 |
| 111334_at | Rtcd1 (RNA terminal phosphate cyclase domain 1) | −4.6 | −3.8 | |
| 95003_at | Polr2k [polymerase (RNA) II (DNA dir.) polypeptide K] | −2.6 | −4.3 | −3.7 |
| 93833_s_at | Hist1h2bc (histone 1, H2bc) | −5.4 | −2.5 | −2.6 |
| 96710_at | H2afv* (H2A histone family, member V) | −1.8 | −1.8 | |
| 108781_at | Smc5l1 (SMC5 structural maint. of chromosomes 5-like 1) | 6.8 | 2.4 | |
| Metabolism | ||||
| 96918_at | Fbp1** (fructose bisphosphatase 1) | 50.0 | 18.7 | |
| 104343_f_at | Pla2g12a (phospholipase A2, group XIIA) | −2.0 | −5.7 | −3.3 |
| 99608_at | Prdx2 (peroxiredoxin 2) | −2.7 | −6.7 | |
| 97819_at | Gsto1 (glutathione S-transferase ω1) | −2.2 | −2.8 | |
| 97402_at | Inmt* (indolethylamine N-methyltransferase) | −3.4 | −2.2 | |
| 95722_at | Glrx1 (glutaredoxin 1) | 1.8 | 2.1 | |
| 160350_at | Gstz1 (glutathione transferase ζ1) | 1.8 | 1.6 | |
| 162718_f_at | Cth (cystathionase) | 2.8 | 1.8 | |
| 93866_s_at | Mgp* (matrix γ-carboxy-glutamate protein) | −2.3 | −3.9 | |
| 100336_s_at | Bglap1,2,-rs1* (bone γ-carboxy-glutamate protein 1, 2, or rs 1) | 3.0 | 3.7 | |
| 100069_at | Cyp2f2* (cytochrome P450, family 2, subfamily f, polypeptide 2) | −3.6 | −3.9 | |
| 96608_at | Phyh (phytanoyl-CoA hydroxylase) | 2.5 | 1.7 | |
| 107220_i_at | Pafah2 (PAF acetylhydrolase 2) | 9.5 | 9.9 | |
| 163212_at | Agpat5 (1-acylglycerol-3-phosphate O-acyltrans-ferase 5) | −4.6 | −5.5 | |
| 103389_at | Aass (aminoadipate-semialdehyde synthase) | 1.6 | 1.8 | |
| 133851_s_at | Osbpl8 (Oxysterol binding protein-like 8) | 1.8 | 1.7 | |
| 96867_at | Azgp1 (α-2-glycoprotein 1, zinc) | −6.4 | −3.8 | |
| Others | ||||
| 94781_at | Hba-a1 (hemoglobin α, adult chain 1) | 2.6 | 1.6 | 2.5 |
| 101869_s_at | Hbb,-b1,-b2* (hemoglobin β chain complex) | 2.6 | 2.7 | |
| 103534_at | Hbb-b2* (hemoglobin, β adult minor chain) | 4.0 | 2.5 | |
| 97825_at | Perp (PERP, TP53 apoptosis effector) | −1.9 | −3.5 | −1.9 |
| 101876_s_at | H2-T10,-T22 (H2, T region loci 9,10,17,22) | −3.1 | −2.4 | |
| 97540_f_at | H2-D1 (H2, D region locus 1) | −2.2 | −4.0 | |
| 102161_f_at | H2-Q2 (H2, Q region locus 2) | −1.7 | −2.2 | |
| 98472_at | H2-T23 (H2, T region locus 23) | 2.5 | 2.1 | |
| 117191_at | Lpo (lactoperoxidase) | 2.0 | 1.9 | |
| 101909_f_at | Mup3 (major urinary protein 3) | −3.4 | −1.8 | |
| 95430_f_at | Spg21 (spastic paraplegia 21) | −4.5 | −3.5 | −5.1 |
| 101362_at | Mapk9 (mitogen-activated protein kinase 9) | 1.8 | 1.6 | |
| 95521_s_at | Zfp68 (zinc finger protein 68) | −2.5 | −3.1 | |
| 102382_at | Arntl (aryl hydrocarbon receptor nuc. transl.-like) | 2.1 | 2.9 | |
| 100032_at | Sp1 (trans-acting transc. factor 1) | 1.8 | 1.7 | |
| 101787_f_at | Ccrn4l (CCR4 carbon catabolite repression 4-like) | −1.7 | −7.2 | |
| 96896_at | Actr2 (ARP2 actin-related protein 2 homolog) | −1.6 | −2.0 | |
Genes are grouped by function. Gene symbols, names, probe sets, and fold changes are listed. Fold change definitions are as in Table 1. Blank fields indicate that the differences between the NOD/scid and B6/scid samples were not significant and/or that the genes were not expressed in particular tissues. Loci in humans and mice that are associated with T1D or T2D and map close to the identified genes or their human counterparts are identified by one asterisk for each locus and are shown in Table 3.
Changes in cell–cell and cell–ECM interactions have also been noted in T2D (22). In that case they were thought to be the consequence of chronic hyperglycemia, which leads to the formation of advanced glycation end products that include posttranslationally modified cell-surface molecules that would be expected to affect cell–cell/ECM interactions.
Altered Protein Folding, Processing, Transport Pathways, and the Induction of ER stress.
The expression of the retinitis pigmentosa GTPase regulator interacting protein 1 (Rpgrip1) was >10-fold lower in all three tissues in NOD/scid mice. Rpgrip1 regulates transport pathways by regulating the retinitis pigmentosa GTPase regulator. It is mapped to murine T1D, T2D, as well as human T2D diabetic loci (Table 3). Analyses of the photoreceptors of mice lacking normal Rpgrip1 expression showed abnormal disk morphogenesis and evidence of ongoing cell death (23). In addition, ≈40 genes with known or putative roles in the targeting of intracellular proteins or in vesicular traffic were differentially expressed across the three tissues. Together these changes suggest that there is protein mislocalization and abnormal vesicular trafficking in NOD/scid autoimmune target tissues.
Table 3.
Candidate genes for diabetes loci
| Locus | Gene symbol | Mouse map | Human map |
|---|---|---|---|
| Murine T1D | |||
| Idd2 | Rcn2 | Chr. 9, 51 cM | 15q23 |
| Idd7 | Cyp2f2 | Chr. 7, 7 cM | 19q13.2 |
| Idd12 | Rpgrip1 | Chr. 14, 20 cM | 14q11 |
| Idd13 | Slc30a4 | Chr. 2, 69 cM | 15q21.1 |
| Idd14 | Fbp1 | Chr. 13, 36 cM | 9q22.3 |
| Idd14 | Ctsl | Chr. 13, 30 cM | 9q21-q22 |
| Idd17 | Ctss | Chr. 3, 43 cM | 1q21 |
| Idd17 | Bglap1,2,-rs1 | Chr. 3, 43 cM | 1q25-q31 |
| Idd19 | Iapp | Chr. 6, 62 cM | 12p12.3-p12.1 |
| Idd19 | Mgp | Chr. 6, 66 cM | 12p13.1-p12.3 |
| Idd19 | Mug1 | Chr. 6, 62 cM | n/a |
| Murine T2D | |||
| Nidd1n | Ube2b | Chr. 11, 30 cM | 5q23-q31 |
| Nidd2n | Rpgrip1 | Chr. 14, 20 cM | 14q11 |
| Nidd4n | H2afv | Chr. 11, 1 cM | 7p13 |
| Nidd5 | Gcg | Chr. 2, 36 cM | 2q36-q37 |
| Human T1D | |||
| IDDM2 | Ins1 | Chr. 19, 49 cM | 11p15.5 |
| IDDM2 | Hbb,-b1,-b2 | Chr. 7, 50 cM | 11p15.5 |
| IDDM4 | Gal | Chr. 19, 2 cM | 11q13.2 |
| IDDM6 | Lman1 | Chr. 18, 38 cM | 18q21.3-q22 |
| Human T2D | |||
| 1q21-q24 | Itlna | Chr. 1, 93 cM | 1q21-q23 |
| 1p36.3-1p36.23 | Ctrc | Chr. 4, 70 cM | 1p36.21 |
| 5q14 | Pam | Chr. 1, 58 cM | 5q14-q21 |
| 6q15-q16 | Slc35a1 | Chr. 4, 15 cM | 6q15 |
| 9q21 | Ctsl | Chr. 13, 30 cM | 9q21-q22 |
| 9q22 | Fbp1 | Chr. 13, 36 cM | 9q22.3 |
| 11q14-q24 | Ctsc | Chr. 7, 46 cM | 11q14.1-q14.3 |
| 11q23-24 | St3gal4 | Chr. 9, 17 cM | 11q23-q24 |
| 12q13-q14 | Slc11a2 | Chr. 15, 60 cM | 12q13 |
| 12q13-q14 | Galgt1 | Chr. 10, 69 cM | 12q13.3 |
| 14q11 | Rpgrip1 | Chr. 14, 20 cM | 14q11 |
| 7p15.3 | Inmt | Chr. 6, 28 cM | 7p15.3-p15.2 |
Disruption of proper protein trafficking is known to induce ER stress. This situation may be compounded by decreased proteolysis in the NOD/scid tissues. Transcripts for a proton ATPase (Atp6v1d), which is necessary for the proper acidification of lysosomes (24), and carboxypeptidase E were also significantly lower in all NOD/scid tissues. Additionally, transcripts for cathepsins S, C, and L and lysosomal peptidases showed decreased expression in several tissues. There were indications of diminished proteosome-dependent degradation as all tissues showed decreased transcripts coding for proteosome subunits and/or enzymes relevant to the ubiquitination of proteins such as the ubiquitin-conjugating enzymes E2b and E2d3 in NOD/scid tissues.
Decreased proteolysis and reduced proper protein trafficking and folding may lead to accelerated accumulation of ill-folded molecules, which have been linked to the production of amyloid aggregates in T2D. Interference with unfolded protein-induced translational control through the elimination of ER kinase–PERK (PKR-like ER kinase) (25) or through the mutation of the serine phosphorylation site of eukaryotic initiation factor 2 (26) has led to an abnormal distension of the ER lumen, the defective trafficking of proinsulin, a reduction in numbers of insulin granules in β cells, a loss of normal glucose homeostasis, and the development of T2D.
Alterations in Metabolism.
Altered phosphorylation of cellular factors is a complication of T2D and is thought to be the consequence of high-glucose-induced accumulation of fructose-6-phosphate that, when shunted into the hexosamine pathway, disturbs the equilibrium of O-linked glycosylation and phosphorylation (22). In NOD/scid mice, transcripts for fructose-1,6-bisphosphatase (Fbp1) were ≈50-fold and ≈19-fold higher than in B6/scid submandibular and lacrimal glands, respectively. Fbp1 catalyzes a committed step in gluconeogenesis: the dephosphorylation of fructose-1,6-bisphosphate to fructose-6-phosphate. High expression of Fbp1 predicts the accumulation of fructose-6-phosphate within cells. Human FBP1 maps to 9q22 and within a 27-Mb region that is linked to abnormal insulin levels in 2-h oral glucose tolerance tests in Pima Indians (27, 28). Murine fbp1 maps into the middle of Idd14. Interestingly, B6 alleles in this locus are found to enhance the incidence of diabetes above NOD levels (29). Additional enzymes involved in fatty acid metabolism, glycolysis, gluconeogenesis, mitochondrial electron transport, calcium homeostasis, and establishment of redox potential were also found to be differentially expressed.
The Exocrine Tissues Are More Active in NOD/scid than in B6/scid Mice.
In general, transcripts encoding exocrine products were expressed at higher levels and suggested that there were altered digestion and immediate antimicrobial responses in NOD/scid tissues. These included chymotrypsin C and salivary protein 2, which were present at ≈20-fold and ≈100-fold greater levels in the pancreata and the submandibular glands of NOD/scid mice, respectively.
Immunologic Implications.
The altered physiology suggested by this study should affect immune responses in multiple ways. In addition to what has been discussed above, the association of glycolysis and fatty acid/lipid metabolism with the outcome of T cell and macrophage activation is well established (30, 31). Vascular changes may allow for greater access to tissues by circulating leukocytes, and the generation of glycosylated ligands that are important in leukocyte migration depends directly on glycosylation enzymes. Abnormal protein processing may alter both the quality and the quantity of self-antigens by inefficient invariant chain editing (e.g., by cathepsin S) and by altered peptide and lipid antigen generation (e.g., by cathepsin L). An implied consequence is the release of immature secretory granules containing misrouted and inappropriately processed membrane and secretory proteins/peptides (32). In this context, several T and B cell epitopes in autoimmune diabetes correspond to the transmembrane regions of β cell proteins, granule components, and the leader and connecting peptides of insulin (33–35). Abnormal protein processing also indirectly affects the development of T cell repertoires (36, 37). Additionally, significant differences in the expression of the MHC molecules D1, Q2, T10/T22, and T23 were found in this comparison. Idd13 is believed to be β2-microglobulin, and it has been suggested that such β2-microglobulin-dependent molecules play a role in T1D (38).
Conclusions
Our survey of three autoimmune target tissues in NOD/scid mice suggests that, although major cellular pathways seem intact, there are significant changes in gene expression patterns that signify the presence of an altered physiology in these mice. These changes include insulin resistance, vascular pathology, metabolic alterations, and accumulated ER protein stress in β cells, which are commonly associated with T2D. Importantly, these changes occur in the absence of high circulating glucose concentrations, which is the end result seen in T2D, and without autoimmune destruction of the target tissues, which is the proximate cause of T1D. Thus, there may be a partial overlap in etiology between T1D and T2D (10, 39, 40), which is masked by hyperglycemia in T2D and autoimmune destruction of islets in T1D.
In fact, epidemiological analyses have shown that T1D may develop more readily in a T2D-predisposed genetic background. For example, a parent with T2D increases the risk for T1D in siblings of a T1D proband, and a high proportion of parents of children with T1D have T2D (reviewed in ref. 10). In the United Kingdom population, up to 20% of T2D patients have anti-islet cell antibodies and/or anti-glutamic acid decarboxylase antibodies. In addition, nondiabetic relatives of T1D patients have exhibited elevated proinsulin, C-peptide, basal insulin, and glucose-stimulated insulin levels, which are commonly associated with T2D (41, 42). Strikingly, the 10–25% prevalence of islet cell antibodies among latent autoimmune diabetes in adult patients is greater than the 0.5% frequency of T1D in the general population (43). Yet, despite these data indicating that metabolic defects that are commonly associated with T2D may also contribute to T1D, analyses of putative T1D-susceptibility loci in familial T2D have shown no evidence of linkage to the majority of these loci, other than to insulin. Our results suggest that the NOD model (and related strains) may be a good candidate to study the interface of these two forms of diabetes and to provide candidate genes and mechanisms for the general physiological deviations of diabetes.
Materials and Methods
Microarray Analysis.
The guidelines for the care and use of animals at Stanford University were followed for all experiments. B6.CB17-Prkdcscid/SzJ (B6/scid) and NOD.CB17-Prkdcscid/J (NOD/scid) female mice were maintained on a 12-h light/dark cycle and fed LabDiet JL Rat and Mouse/Auto 6F ovals (catalog no. 5K67, PMI Nutrition International). Mice were killed by CO2 asphyxiation. Lacrimal glands and pancreata were immediately removed and homogenized in TRIzol reagent (Invitrogen). Submandibular glands were separated away from sublingual glands and connective and adipose tissue of the salivary glands before homogenization. Total RNA was isolated, and cDNA and biotinylated cRNA were prepared and hybridized to Murine Genome U74v2 microarrays per the manufacturer’s instructions (Affymetrix) without additional amplification steps.
Microarray data from each sample were initially scaled to a mean intensity value of 2,500 by using mas (Microarray Suite) software, version 5.0 (Affymetrix). Probe sets were determined to be present, absent, or marginal based on default parameters. Thereafter, all data were transferred to spreadsheet software for cube-root normalization and differential expression analysis by the sam (significance analysis of microarrays) statistical package (44).
In sam, each gene is assigned a score on the basis of its change in expression relative to the standard deviation of repeated measurements for that gene. In this study, relative differences of gene expression levels were computed from permutations of the hybridization of five independent NOD/scid and five independent B6/scid samples. The data were treated as two-class unpaired data. An observed relative difference, d(i), was calculated for each gene based on the ratio of the change in gene expression to the standard deviation in the data for that gene. An expected relative difference, dE(i), was also calculated for each gene. Genes whose d(i) and dE(i) values were displaced from the d(i) = dE(i) diagonal by a distance greater than a threshold, Δ, of 1.5 and that showed an increase or decrease of at least 1.5-fold between the NOD/scid and B6/scid samples were called significantly changed. Under these conditions, the false discovery rate, defined as the percentage of falsely identified significant genes to the genes called significant, was <5%.
Insulin and Glucose Tolerance Tests.
Six-week-old NOD/scid and B6/scid female mice were used for the studies. For insulin challenge, mice were injected i.p. with 0.75 or 2 units/kg Humulin R (Eli Lilly). For glucose challenge, mice were first fasted overnight for 16 h and then injected i.p. with 2 g/kg glucose. To assess insulin release, mice were injected i.p. with 3 g/kg glucose. At each time point mice were warmed for 3 min under an incandescent lamp to facilitate blood collection. Approximately 30 μl of tail vein blood was drawn and centrifuged at 14,000 × g for 5 min at room temperature to separate serum from cells. Insulin content in serum was measured by using a mouse insulin ELISA kit (ALPCO Diagnostics, Windham, NH). Blood glucose measurements were made by using a Freestyle blood glucose meter (Therasense, Alameda, CA).
Supplementary Material
Acknowledgments
We thank Drs. John A. Todd and Hironori Ueda for critically reviewing the manuscript and for suggestions and Dr. Sara Michie for consultation. R.J.C. was a Ford Foundation Predoctoral Fellow. R.J.C and Y.K. were partially supported by National Institutes of Health Training Grant T32 AI07290. This work was supported by National Institutes of Health Grant P01 DK53001 (to Y.-h.C.).
Abbreviations
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- NOD
nonobese diabetic
- scid
severe combined immunodeficient
- ECM
extracellular matrix
- ER
endoplasmic reticulum
- Fbp1
fructose-1,6-bisphosphatase
- B6
C57BL/6.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE4953).
See Commentary on page 12217.
References
- 1.Serreze D. V., Leiter E. H. Curr. Opin. Immunol. 1994;6:900–906. doi: 10.1016/0952-7915(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 2.Christianson S. W., Shultz L. D., Leiter E. H. Diabetes. 1993;42:44–55. doi: 10.2337/diab.42.1.44. [DOI] [PubMed] [Google Scholar]
- 3.Anderson M. S., Bluestone J. A. Annu. Rev. Immunol. 2005;23:447–485. doi: 10.1146/annurev.immunol.23.021704.115643. [DOI] [PubMed] [Google Scholar]
- 4.Homo-Delarche F., Drexhage H. A. Trends Immunol. 2004;25:222–229. doi: 10.1016/j.it.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Homo-Delarche F. Diabetes Metab. 1997;23:181–194. [PubMed] [Google Scholar]
- 6.Humphreys-Beher M. G., Peck A. B. Arch. Oral Biol. 1999;44(Suppl. 1):S21–S25. doi: 10.1016/s0003-9969(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 7.Hunger R. E., Muller S., Laissue J. A., Hess M. W., Carnaud C., Garcia I., Mueller C. J. Clin. Invest. 1996;98:954–961. doi: 10.1172/JCI118879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson C. P., Yamachika S., Alford C. E., Cooper C., Pichardo E. L., Shah N., Peck A. B., Humphreys-Beher M. G. Proc. Natl. Acad. Sci. USA. 1997;94:5767–5771. doi: 10.1073/pnas.94.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda H., Ikegami H., Yamato E., Fu J., Fukuda M., Shen G., Kawaguchi Y., Takekawa K., Fujioka Y., Fujisawa T., et al. Diabetologia. 1995;38:503–508. doi: 10.1007/BF00400717. [DOI] [PubMed] [Google Scholar]
- 10.Homo-Delarche F. Diabetes Metab. 1997;23:473–505. [PubMed] [Google Scholar]
- 11.Yamano S., Atkinson J. C., Baum B. J., Fox P. C. Clin. Immunol. 1999;92:265–275. doi: 10.1006/clim.1999.4759. [DOI] [PubMed] [Google Scholar]
- 12.Pelegri C., Rosmalen J. G., Durant S., Throsby M., Alves V., Coulaud J., Esling A., Pleau J. M., Drexhage H. A., Homo-Delarche F. Mol. Med. 2001;7:311–319. [PMC free article] [PubMed] [Google Scholar]
- 13.Goren H. J., Kulkarni R. N., Kahn C. R. Endocrinology. 2004;145:3307–3323. doi: 10.1210/en.2003-1400. [DOI] [PubMed] [Google Scholar]
- 14.Withers D. J., Gutierrez J. S., Towery H., Burks D. J., Ren J. M., Previs S., Zhang Y., Bernal D., Pons S., Shulman G. I., Bonner-Weir S., White M. F. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 15.Amrani A., Jafarian-Tehrani M., Mormede P., Durant S., Pleau J. M., Haour F., Dardenne M., Homo-Delarche F. J. Endocrinol. 1996;148:139–148. doi: 10.1677/joe.0.1480139. [DOI] [PubMed] [Google Scholar]
- 16.Dickinson D. P., Gross K. W., Piccini N., Wilson C. M. Genetics. 1984;108:651–667. doi: 10.1093/genetics/108.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piccini N., Knopf J. L., Gross K. W. Cell. 1982;30:205–213. doi: 10.1016/0092-8674(82)90026-5. [DOI] [PubMed] [Google Scholar]
- 18.Wilson C. M., Cherry M., Taylor B. A., Wilson J. D. Biochem. Genet. 1981;19:509–523. doi: 10.1007/BF00484623. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu S., Kiyosawa H., Yoshiki A., Okazaki Y., Yoshino M., Tomaru Y., Watanabe S., Muramatsu M., Kusakabe M., Hayashizaki Y. Mamm. Genome. 2002;13:293–298. doi: 10.1007/s00335-001-2134-7. [DOI] [PubMed] [Google Scholar]
- 20.Zammit D. J., Berzins S. P., Gill J. W., Randle-Barrett E. S., Barnett L., Koentgen F., Lambert G. W., Harvey R. P., Boyd R. L., Classon B. J. Mol. Cell. Biol. 2002;22:946–952. doi: 10.1128/MCB.22.3.946-952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geering K. Am. J. Physiol. 2006;290:F241–F250. doi: 10.1152/ajprenal.00126.2005. [DOI] [PubMed] [Google Scholar]
- 22.Brownlee M. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y., Hong D. H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A., Li T. Proc. Natl. Acad. Sci. USA. 2003;100:3965–3970. doi: 10.1073/pnas.0637349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens T. H., Forgac M. Annu. Rev. Cell Dev. Biol. 1997;13:779–808. doi: 10.1146/annurev.cellbio.13.1.779. [DOI] [PubMed] [Google Scholar]
- 25.Sonenberg N., Newgard C. B. Science. 2001;293:818–819. doi: 10.1126/science.1062937. [DOI] [PubMed] [Google Scholar]
- 26.Scheuner D., Mierde D. V., Song B., Flamez D., Creemers J. W., Tsukamoto K., Ribick M., Schuit F. C., Kaufman R. J. Nat. Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 27.Kim S. H., Ma X., Klupa T., Powers C., Pezzolesi M., Warram J. H., Rich S. S., Krolewski A. S., Doria A. Diabetes. 2003;52:2182–2186. doi: 10.2337/diabetes.52.8.2182. [DOI] [PubMed] [Google Scholar]
- 28.Pratley R. E., Thompson D. B., Prochazka M., Baier L., Mott D., Ravussin E., Sakul H., Ehm M. G., Burns D. K., Foroud T., et al. J. Clin. Invest. 1998;101:1757–1764. doi: 10.1172/JCI1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brodnicki T. C., Quirk F., Morahan G. Diabetes. 2003;52:218–222. doi: 10.2337/diabetes.52.1.218. [DOI] [PubMed] [Google Scholar]
- 30.Fox C. J., Hammerman P. S., Thompson C. B. Nat. Rev. Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 31.Castrillo A., Tontonoz P. Annu. Rev. Cell Dev. Biol. 2004;20:455–480. doi: 10.1146/annurev.cellbio.20.012103.134432. [DOI] [PubMed] [Google Scholar]
- 32.Kuliawat R., Arvan P. J. Cell Biol. 1994;126:77–86. doi: 10.1083/jcb.126.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dotta F., Di Mario U. Acta Pathol. Microbiol. Immunol. Scand. 1996;104:769–774. doi: 10.1111/j.1699-0463.1996.tb04941.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama M., Abiru N., Moriyama H., Babaya N., Liu E., Miao D., Yu L., Wegmann D. R., Hutton J. C., Elliott J. F., Eisenbarth G. S. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kent S. C., Chen Y., Bregoli L., Clemmings S. M., Kenyon N. S., Ricordi C., Hering B. J., Hafler D. A. Nature. 2005;435:224–228. doi: 10.1038/nature03625. [DOI] [PubMed] [Google Scholar]
- 36.Maehr R., Mintern J. D., Herman A. E., Lennon-Dumenil A. M., Mathis D., Benoist C., Ploegh H. L. J. Clin. Invest. 2005;115:2934–2943. doi: 10.1172/JCI25485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honey K., Benlagha K., Beers C., Forbush K., Teyton L., Kleijmeer M. J., Rudensky A. Y., Bendelac A. Nat. Immunol. 2002;3:1069–1074. doi: 10.1038/ni844. [DOI] [PubMed] [Google Scholar]
- 38.Hamilton-Williams E. E., Serreze D. V., Charlton B., Johnson E. A., Marron M. P., Mullbacher A., Slattery R. M. Proc. Natl. Acad. Sci. USA. 2001;98:11533–11538. doi: 10.1073/pnas.191383798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDevitt H. O. Diabetes. 2005;54(Suppl. 2):S4–S10. doi: 10.2337/diabetes.54.suppl_2.s4. [DOI] [PubMed] [Google Scholar]
- 40.Ikegami H., Fujisawa T., Ogihara T. ILAR J. 2004;45:268–277. doi: 10.1093/ilar.45.3.268. [DOI] [PubMed] [Google Scholar]
- 41.Heaton D. A., Millward B. A., Gray P., Tun Y., Hales C. N., Pyke D. A., Leslie R. D. Br. Med. J. 1987;294:145–146. doi: 10.1136/bmj.294.6565.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Villar C., Conget I., Casamitjana R., Vidal J., Manzanares J. M., Gomis R. Diabetes Res. Clin. Pract. 1997;37:145–148. doi: 10.1016/s0168-8227(97)00055-7. [DOI] [PubMed] [Google Scholar]
- 43.Palmer J. P., Hampe C. S., Chiu H., Goel A., Brooks-Worrell B. M. Diabetes. 2005;54(Suppl. 2):S62–S67. doi: 10.2337/diabetes.54.suppl_2.s62. [DOI] [PubMed] [Google Scholar]
- 44.Tusher V. G., Tibshirani R., Chu G. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.