Abstract
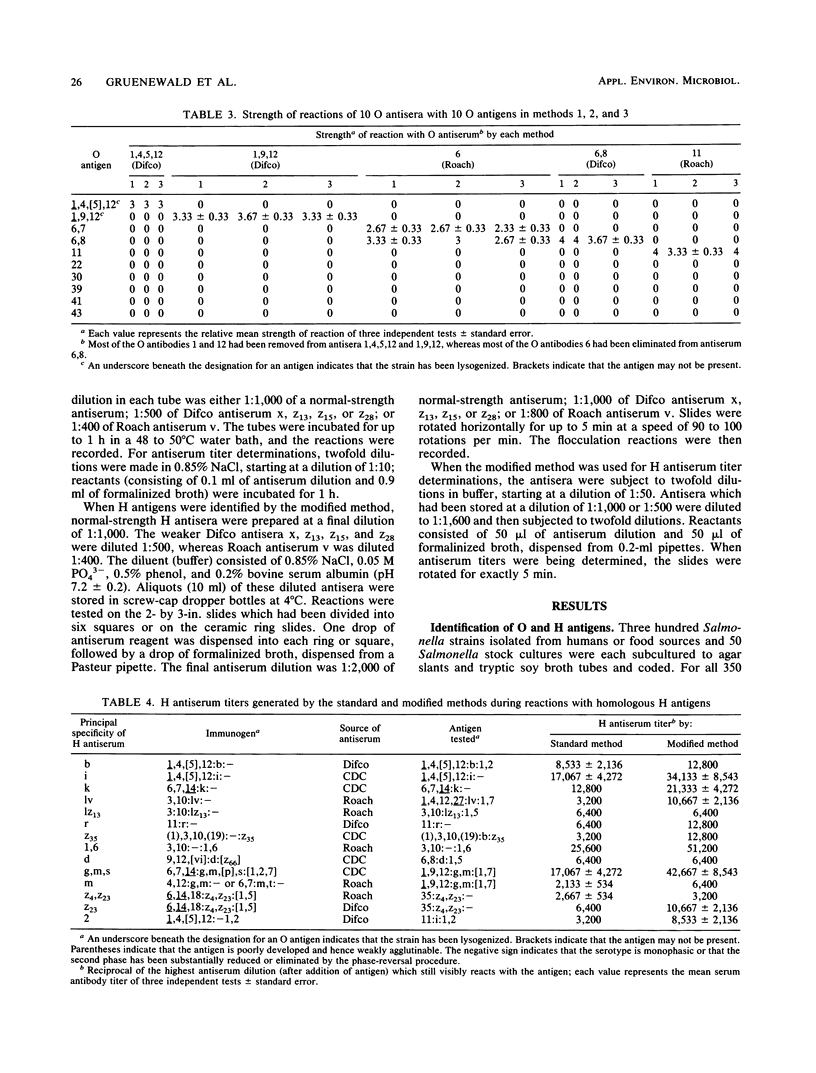
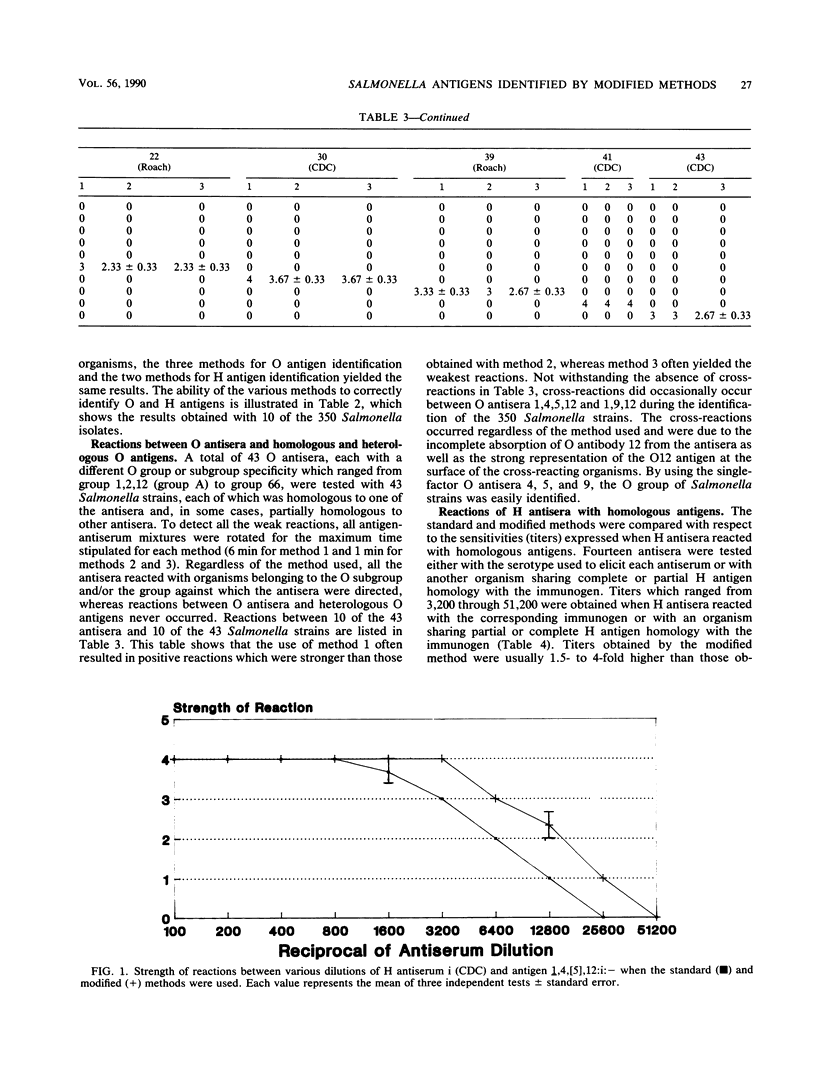
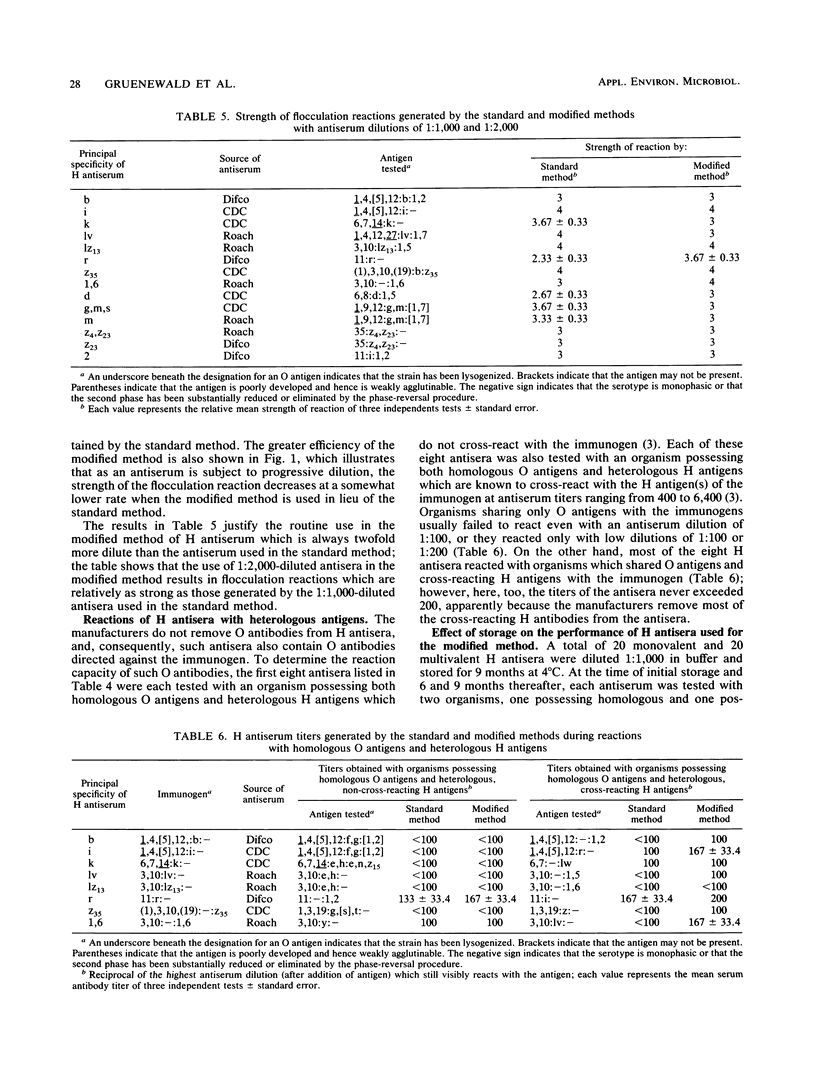
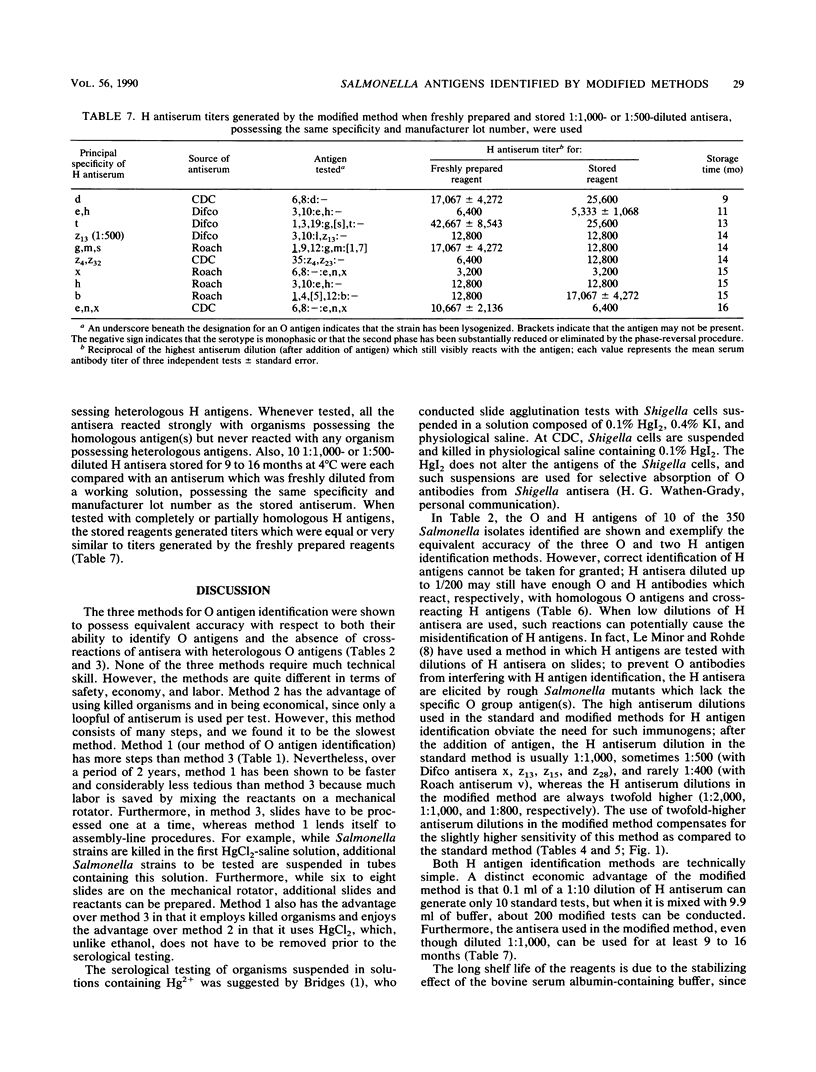
This report describes two modified methods for the identification of Salmonella somatic (O) and flagellar (H) antigens. Over a period of 2 years, both modified methods were found to be approximately three times less labor intensive than the standard methods while requiring no more technical skill. The modified methods were as accurate as the standard methods in identifying the O and H antigens of 350 Salmonella isolates. Furthermore, 43 O antisera reacted exclusively with organisms possessing homologous O antigens when the modified and two standard methods were used. At the antiserum dilutions used for H antigen identification, H antisera did not react with O antigens or heterologous H antigens by either the modified or the standard method. Compared with the standard method for H antigen identification, the modified method was approximately 20 times more economical with respect to antisera and usually generated a 1.5- to 4-fold higher titer. Since the antisera stored for use in the modified method for H antigen identification were usually 100-fold more dilute than the antisera stored for the standard method, an antibody-stabilizing buffer was incorporated in the diluted antisera, allowing these reagents to be used for at least 9 to 16 months.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRIDGES R. F. The dysentery reference laboratory. Br Med Bull. 1951;7(3):200–203. doi: 10.1093/oxfordjournals.bmb.a073834. [DOI] [PubMed] [Google Scholar]
- Cohen J. O., Britt L. E., Harrell W. K. Identification of Salmonella O antigens by coagglutination. J Clin Microbiol. 1984 May;19(5):576–578. doi: 10.1128/jcm.19.5.576-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer J. J., 3rd Formalinized bacterial "antigens" as a potential infection hazard. J Clin Microbiol. 1975 Oct;2(4):359–360. doi: 10.1128/jcm.2.4.359-360.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenewald R., Stollar B. D. The role of antigenic determinants in the control of IgM and IgG antibody responses to denatured DNA. J Immunol. 1973 Jul;111(1):106–113. [PubMed] [Google Scholar]
- Guinée P. A., Jansen W. H., Maas H. M. Mechanized procedures for the serology of Salmonella. Zentralbl Bakteriol Mikrobiol Hyg A. 1983 Sep;255(2-3):258–264. [PubMed] [Google Scholar]
- Nikitin V. M., Roshchin Iu N., Kotich A. A. Novaia forma diagnosticheskikh preparatov--immunoindikatornye karandashi. Lab Delo. 1986;(7):438–440. [PubMed] [Google Scholar]
- Shipp C. R., Rowe B. A mechanised microtechnique for salmonella serotyping. J Clin Pathol. 1980 Jun;33(6):595–597. doi: 10.1136/jcp.33.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WASSERMAN E., LEVINE L. Quantitative micro-complement fixation and its use in the study of antigenic structure by specific antigen-antibody inhibition. J Immunol. 1961 Sep;87:290–295. [PubMed] [Google Scholar]


