Abstract
To detect interactions between proteins of vaccinia virus, we carried out a comprehensive two-hybrid analysis to assay every pairwise combination. We constructed an array of yeast transformants that contained each of the 266 predicted viral ORFs as Gal4 activation domain hybrid proteins. The array was individually mated to transformants containing each ORF as a Gal4–DNA-binding domain hybrid, and diploids expressing the two-hybrid reporter gene were identified. Of the ≈70,000 combinations, we found 37 protein–protein interactions, including 28 that were previously unknown. In some cases, e.g., late transcription factors, both proteins were known to have related roles although there was no prior evidence of physical associations. For some other interactions, neither protein had a known role. In the majority of cases, however, one of the interacting proteins was known to be involved in DNA replication, transcription, virion structure, or host evasion, thereby providing a clue to the role of the other uncharacterized protein in a specific process.
Poxviruses are large, complex, double-stranded DNA viruses that replicate in the cytoplasm of infected cells (1). Vaccinia virus, the best-characterized member of this large family, was extensively used as the smallpox vaccine, has gained popularity as a mammalian expression vector, and is being tested as a recombinant vaccine against cancer and infectious diseases (2). Vaccinia virus has a genome of approximately 190 kbp and can potentially express more than 200 proteins, allowing an exceptional degree of independence from the host (3). Virus-encoded proteins involved in transcription include a multicomponent DNA-dependent RNA polymerase, an assortment of transcription factors, and enzymes that cap, methylate, and polyadenylylate mRNA (4). Of the eight virus-encoded proteins that have been implicated in DNA replication, four are directly involved in DNA polymerization, and others (such as a type I DNA topoisomerase, a single-stranded DNA-binding protein, a DNA ligase, and a DNA–RNA helicase) have other roles (5). Additional viral proteins are needed to maintain adequate levels of deoxyribonucleotides for DNA replication, including a thymidine kinase, a thymidylate kinase, a deoxyribonucleotide reductase, and a deoxyuridine triphosphatase. At least 30 proteins form the core and membrane components of virus particles (6, 7). Other viral proteins interact with host components to facilitate virus dissemination, prevent apoptosis, and attenuate immune responses (8, 9). The study of vaccinia virus thus provides important information that will help us to understand the nature of more pathogenic members of the family, such as the agents of smallpox, monkeypox, and molluscum contagiosum (10), as well as insights into many areas of molecular and cellular biology and immunology.
Although 10 years have passed since the genome of vaccinia virus was sequenced (3), the roles of about half of the genes remain entirely unknown. A similar situation exists for other large DNA viruses, including members of the herpesvirus family. To increase our understanding of the poxvirus life cycle and to evaluate an approach that would be generally applicable to other large viruses, we initiated a genome-wide yeast two-hybrid analysis to identify vaccinia virus protein–protein interactions. Each of the ≈70,000 potential pairwise combinations of proteins was assayed, identifying putative interactions among both characterized viral proteins and those of unknown function.
Materials and Methods
Construction of pOBD2-20.
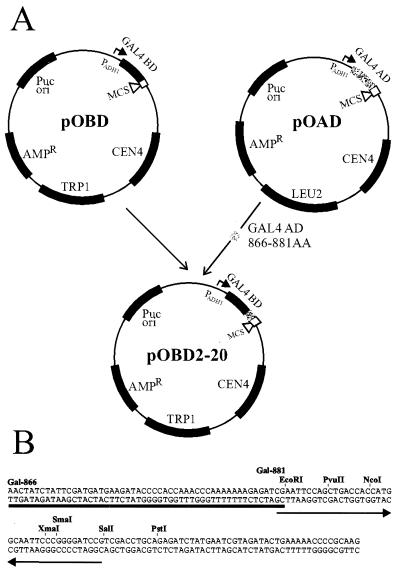
A new DNA-binding domain vector was generated containing Gal4 residues 1 to 147 and a small portion of the activation domain plasmid pOAD (11) that encodes Gal4 residues 866 to 881 along with a shared multicloning site (Fig. 1A). PCR was used to amplify the Gal4 DNA-binding domain from the plasmid pCPL2 (12) by using primer P1 5′-(GTTCTCGTTCCCTTTCTTCCTTGT)-3′ located in the ADH1 promoter region and primer P2 5′-CGATCTCTTTTT-TTGGGTTTGGTGGGGTATCTTCATCATCGAATAGAT-AGTT(CGATACAGTCAACTGTCTTTGACC)-3′ corresponding to residues 866–881 of the Gal4 activation domain. The primer sequences shown in parentheses are complementary to those present in pCPL2, and the underlined bases are an added sequence. The additional sequence is complementary to sequences in pOAD. In a separate PCR, a portion of the multicloning site and ADH1 terminator sequence was amplified from pOAD by using primer P3 5′-AACTATCTATTCGATGATGAAGATACCCCACCAAACCCAAAAAAAGAGATCGAATTCCAGCTGACCACCATGGCAA-3′ and primer P4 5′-AAATTTCTGGCAAGGTAGACAAGC-3′. These two regions were joined in a subsequent PCR amplification using primers P1 and P4 and a mixture of the two initial PCR products as a template. This second product was digested with HindIII and ligated into HindIII-cut and calf intestinal phosphatase-treated pCPL2. The resultant vector, pOBD2-20, was confirmed by sequence analysis. The inserted portion contains a region common to both pOAD and pOBD2-20 (Fig. 1B), and thus a set of PCR products with appropriate 5′ and 3′ sequences can be cloned by homologous recombination into both vectors.
Figure 1.
Scheme for construction of pOBD2-20. (A) A small portion of the Gal4 activation domain was PCR amplified and transferred to the Gal4–DNA-binding domain vector pOBD1 (11) to create the new vector pOBD2-20. (B) Common sequence flanking the multicloning site of pOAD1 and pOBD2-20. A portion of this shared sequence was added to each vaccinia ORF by PCR amplification to allow recombinational cloning of these ORFs into pOBD2-20.
Selection of Vaccinia Virus ORFs.
The map program from the Wisconsin Package version 10.0 [Genetics Computer Group (GCG), Madison, WI] was used to identify potential coding sequences in all six reading frames of 65 or more residues in the genome of the WR strain of vaccinia virus. Sequence files representing the coding regions for each of these ORFs were created. For each of these sequences, a list was constructed containing 15 potential forward primers consisting of the 5′ 18, 19, … , or 32 nucleotides of coding region starting with the predicted ATG, and 15 potential reverse primers containing the reverse complement of the last 18, 19, … , or 32 nucleotides ending with the predicted stop codons. Using the GCG prime program, we selected the most promising primer pairs on the basis of limited folding parameters and with optimal annealing temperatures of ≈55°C. Twenty ORFs whose primer sets did not meet these criteria were further analyzed by using oligo 4.0 primer analysis software (National Biosciences, Plymouth, MN), and an adjustment of the relative position of the start or stop codon within the primer set was made to make suitable primers. To allow for manipulation of the vaccinia primer set en masse, we added a common sequence (5′-CGAATTCCAGCTGACCACC-3′) at the 5′ of each of the forward primers, and a common sequence (5′-CGGATCCCCGGGAATTGC-3′) at the 5′ of each of the reverse primers. Primers were synthesized by Research Genetics (Huntsville, AL).
Preparation of Viral DNA.
Vaccinia virus was purified by sucrose gradient sedimentation, and DNA was extracted as described (13).
Amplification of ORFs.
Each vaccinia virus ORF was amplified from viral genomic DNA in HotStart50 tubes (Life Technologies) essentially as described by the manufacturer. Briefly, a 50-μl reaction mix was prepared containing 0.3 μM each of a forward and reverse primer pair, 250 μM each deoxynucleotide, 0.2 ng of vaccinia DNA, 2.6 units of Expand High Fidelity (Boehringer Mannheim), and 1× Expand buffer. The primary PCRs were carried out for 20 cycles, and 0.2 μl of each resultant product was then reamplified for 15 cycles with primers P5 5′-AGATACCCCACCAAACCCAAAAAAAGAGAT(CGAATTCCAGCTGACCACCATG)-3′ and P6 5′-GGGTTTTTCAGTATCTACGATTCATAGATCTCTGCAGGTCGA(CGGATCCCCGGGAATTGC)-3′ (Fig. 1B). The products were analyzed by electrophoresis on a 0.8% agarose gel and ethidium bromide staining. The size of each ORF was estimated by comparison with DNA standards. Two ORFs that failed to amplify were successfully amplified on another attempt.
Generation of the Activation Domain Array.
The reamplified PCR products were cotransformed with linearized pOAD into PJ69-4A, and with linearized pOBD2-20 into PJ69-4α, as described (11, 14). Transformants were grown overnight in yeast extract/peptone/dextrose (YEPD) medium (15) and stored in 15% glycerol at −70°C. Transformants were recovered by regrowth in YEPD for 6 h, followed by selection on 35-mm synthetic plates (15) lacking either leucine (pOAD transformants) or tryptophan (pOBD2-20 transformants).
Each activation domain transformation was represented by four individual clonal isolates in the array. Additionally, four isolates of transformants carrying activation domain hybrids of either the yeast Mec3 or Rad17 proteins, or human lamin, as well as four transformants of the vector pOAD, were used as control positions in the array. Colonies were transferred to 2 ml of YEPD for overnight growth in 96-deep-well dishes (USA/Scientific, Ocala, FL). Cultures in PJ69-4A were then transferred to a 384-well dish and stamped onto three Omnitrays (Nalgene Nunc) containing synthetic medium lacking leucine. The transfers were done with a Biomek 2000 Laboratory Automation Workstation (Beckman Coulter) equipped with a high density replicating tool. Thus, the vaccinia virus array consisted of 1,076 colonies on three Omnitrays. One ORF, K7R (a protein of unknown function), appeared to be toxic, as yeast containing this gene were not able to grow on selective medium.
Two-Hybrid Screens.
For each ORF, two pOBD2-20 transformants in PJ69-4α were selected on synthetic plates lacking tryptophan and grown for 48 h in 2.5 ml of YEPD medium. The two cultures were pooled and mated on YEPD plates to the PJ69-4A activation domain array by using a 384-pin high density replicating tool. After 3 days of growth at 30°C, diploids containing both an activation domain and a DNA-binding domain plasmid were selected by transfer to synthetic medium lacking leucine and tryptophan and incubated 3 days more. Two-hybrid selection was performed by replicating the diploid array onto medium lacking leucine, tryptophan, and histidine and supplemented with 3 mM 3-amino-1,2,4-triazole. Growth was scored after 1 and 2 weeks of incubation at 30°C.
Results and Discussion
Construction of an Array of Vaccinia Virus Proteins.
Our strategy was to test each pairwise combination of vaccinia virus ORFs in the yeast two-hybrid assay by the use of a protein array (16). The array consisted of a set of yeast transformants, each expressing one vaccinia virus ORF as a hybrid protein with the Gal4 activation domain. This array was mated to yeast transformants of the opposite mating type carrying one of the vaccinia virus ORFs as a hybrid protein with the Gal4 DNA-binding domain, and the diploids were placed onto medium selective for the two-hybrid reporter gene. In this fashion, a single protein could be tested for interaction with every vaccinia virus protein in the array. By generating the complete sets of activation and DNA-binding domain hybrids with vaccinia proteins, we could carry out the mating and selection experiment ≈260 times to assay all of the ≈70,000 combinations.
Using locally written Unix-based scripts for automation of the GCG package [Wisconsin Package Version 10.0, Genetics Computer Group (GCG)], we analyzed the coding sequence of vaccinia virus (WR) for ORFs that were 65 codons or longer. Although fewer than 200 were considered most likely to be expressed, we used a larger set of 266 for the sake of completeness. Each ORF was compared with the set of ORFs for vaccinia virus (Copenhagen strain) (3). ORFs that did not align with existing Copenhagen ORFs were designated simply by their position on the vaccinia virus WR genome, for example F-181498–181845. We then selected an optimal PCR primer pair for each ORF based on the following criteria. The forward primer contained the predicted initiator ATG and 15–29 additional nucleotides of coding sequence, to yield an annealing temperature of 55°C. Each forward primer also contained 19 common nucleotides at its 5′ end to allow reamplification (see Materials and Methods). The reverse primer for each ORF contained the reverse complement of both the predicted termination codon and the preceding 15–29 nucleotides at the end of the reading frame, to have a compatible annealing temperature. Each of these reverse primers also contained 18 common nucleotides. Additionally, primers were selected to minimize sequences that would allow the generation of primer dimers.
Each ORF was amplified individually by PCR through the use of the specific primer pairs. These initial PCR products were then individually reamplified with a common 52-base forward primer and a common 60-base reverse primer. This second round of PCR converted the vaccinia virus ORFs into DNA fragments with common flanking sequences that allow efficient homologous recombination into linearized yeast vectors (17). Cotransformation of these fragments with the vector pOAD (12) into yeast strain PJ69-4A (14) generated a set of 266 activation domain hybrid proteins. The same set of fragments was transformed with the vector pOBD2-20 into yeast strain PJ69-4α (14) to generate a set of DNA-binding domain hybrids. Analysis of recovered plasmids by restriction enzyme analysis indicated that successful insertion of the ORF fragment had occurred in greater than 94% of the transformations, for a representative selection of 96 different ORFs (data not shown).
Identification of Vaccinia Virus Protein–Protein Interactions.
Two-hybrid screens generate significant numbers of false positives, which are not reproducible in a duplicate screen. This random generation of histidine-positive colonies can result from rearrangements and deletions of the DNA-binding domain plasmid, recombinational events between the DNA-binding and activation domain plasmids, and genomic rearrangements of the host strain. To enable the rapid detection of reproducible two-hybrid interactions, we constructed the vaccinia virus array with four separate yeast transformants, corresponding to each activation domain hybrid protein. Thus, the array consisted of 1,064 colonies of these transformants, plus controls, which required three microtiter-sized plates of 384-colony capacity. The array of activation domain transformants was screened against each viral DNA-binding domain transformant. Growth on plates lacking histidine, selective for expression of the GAL1–HIS3 reporter gene, was observed either as a cluster of sister colonies, or as dispersed single colonies that represent false positives. In 20 cases, only 2 of the 4 array positions of a single ORF scored as positive, and in 19 of these 20 the 2 positions were not reproducible in a repetition of the screen. Statistical analysis of the data, based on an average 3.25 random positives per plate and a total of ≈62,000 pairwise assays, predicts 29 events in which 2 of the 4 potential array positions of an ORF would score positive by chance alone, not statistically different from the number observed (Z score = −1.75). As a consequence of this background of nonreproducible positives, only protein combinations that resulted in histidine prototrophy for three or four independent colonies were scored as protein interactions (we would predict 0.17 event in which three of the four positions would occur by chance alone), along with the single pair of A21L with A6L, which was reproducibly positive in two of the four colonies.
Twenty-five proteins (A21L, A26L, A35R, A40R, A41L, A42R, A4L, B ORF A, B3R, B4R, C19L, C7L, C9L, D13L, D3R, E ORF C, E11L, F-181498–181845, F14L, F1L, G2R, K5L, O2L, K7R, YHR1_VACCV) were found to be strong activators when fused to the Gal4 DNA-binding domain, and thus were refractory to analysis by the two-hybrid assay. However, they could be tested unidirectionally as fusions with the Gal4-activation domain. Some of these strong activators may be unrecognized viral transcription factors. Furthermore, any protein that resulted in histidine-positive growth with any of the four controls (the yeast proteins Mec3 and Rad17, human lamin, and the empty vector pOAD) was classified as a false positive and removed from the set of positive interactions.
The testing of 266 vaccinia DNA-binding domain constructs resulted in the scoring of 37 potential interactions (Table 1). The interactions can be grouped based on the participating proteins into five broad categories: DNA replication, transcription, virion structure/morphogenesis, virus–host interactions, and function unknown. Whereas the set of interactions includes a number of previously known combinations as well as homodimers (or higher order multimers), as with any two-hybrid data, the putative new interactions need to be confirmed by other biological or biochemical experiments. Nevertheless, it should be realized that the likelihood of a nonsignificant protein interaction occurring by chance is much lower when screening 1 viral gene against 200 other viral genes compared with the more common screening of a eukaryotic library of about 100,000 genes.
Table 1.
Two-hybrid interactions of vaccinia virus proteins
| BD | AD | ||
|---|---|---|---|
| DNA replication | |||
| A20R | Putative polymerase processivity factor | H5R | Late transcription factor 4 |
| A20R | Putative polymerase processivity factor | D5R | Putative DNA replication factor |
| A20R | Putative polymerase processivity factor | D4R | Uracil DNA glycosylase |
| F2L*† | dUTPase | F2L | dUTPase |
| F4L†‡ | Ribonucleotide reductase small subunit | I4L | Ribonucleotide reductase large subunit |
| F4L*† | Ribonucleotide reductase small subunit | F4L | Ribonucleotide reductase small subunit |
| I3L* | Single-stranded DNA-binding protein | I3L | Single-stranded DNA-binding protein |
| I4L*† | Ribonucleotide reductase large subunit | I4L | Ribonucleotide reductase large subunit |
| J2R*† | Thymidine kinase | J2R | Thymidine kinase |
| Transcription | |||
| A ORF D | Hypothetical 9.8-kDa protein | A23R | Intermediate transcription factor 3 large subunit |
| A1L | Late transcription factor 2 | G8R | Late transcription factor 1 |
| A23R† | Intermediate transcription factor 3 large subunit | A8R | Intermediate transcription factor 3 small subunit |
| A49R | Hypothetical 18.8-kDa protein | H5R | Late transcription factor 4 |
| D7R | RNA polymerase subunit RPO 18 | A32L | Putative DNA-packaging protein, morphogenesis |
| F8L‡ | Hypothetical 7.8-kDa protein | L4R | DNA binding, helicase stimulation, virion transcription |
| G2R†‡ | Transcription elongation factor | H5R | Late transcription factor 4 |
| G8R* | Late transcription factor 1 | G8R | Late transcription factor 1 |
| H5R | Late transcription factor 4 | B1R | Protein kinase 1 |
| Virion structure | |||
| A10L* | Major core protein P4A | A10L | Major core protein P4A |
| A12L‡ | Virion protein | A19L | Hypothetical 8.3-kDa protein |
| A14L | Phosphorylated membrane protein | A40R | Hypothetical 19.3-kDa NK cell receptor homolog |
| A32L | Putative DNA-packaging protein, morphogenesis | A11R | Hypothetical 36.1-kDa protein |
| A45R | Superoxide dismutase (Cu-Zn) homolog | A4L | Virion protein, between core and membrane |
| J1R | Virion protein | A45R | Superoxide dismutase-like (Cu-Zn) |
| J1R* | Virion protein | J1R | Virion protein |
| Virus–host interactions | |||
| A26L | Cowpox A-type inclusion protein homolog | A42R | Profilin homolog |
| E3L*† | Double-stranded RNA-binding protein, IFN resistance | E3L | Double-stranded RNA-binding protein, IFN resistance |
| K1L† | Host range factor | C10L | Hypothetical 38.5-kDa protein |
| E7R | Soluble, myristylated, nonessential | A39R | Semaphorin, cytokine induction |
| Unknown function | |||
| A21L | Hypothetical 13.6-kDa protein | A6L | Hypothetical 43.1-kDa protein |
| A22R* | Hypothetical 20.7-kDa protein | A22R | Hypothetical 20.7-kDa protein |
| A45R* | Superoxide dismutase (Cu-Zn) homolog | A45R | Superoxide dismutase (Cu-Zn) homolog |
| A51R* | Hypothetical 31.7-kDa nonessential protein | A51R | Hypothetical 31.7-kDa nonessential protein |
| E8R | Hypothetical 31.9-kDa protein | A51R | Hypothetical 31.7-kDa nonessential protein |
| F15L | Hypothetical 18.6-kDa protein | D9R | MutT family member |
| 12L | Hypothetical 35.8-kDa protein | A40R | Hypothetical 19.3-kDa NK cell receptor homolog |
| Unnamed‡ | Hypothetical 8.9-kDa protein (ORF position R-111049-111279) | E10R | Hypothetical 10.8-kDa protein |
*Self-interacting proteins.
†Previously observed interactions.
‡Reciprocal interactions observed in both directions.
DNA Replication.
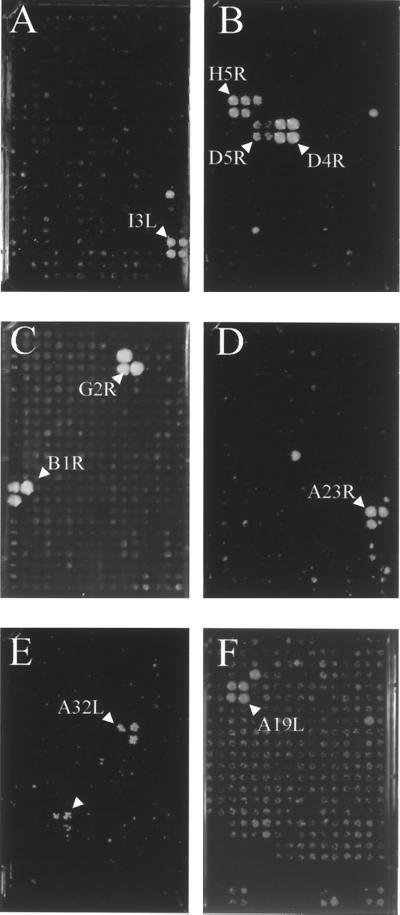
Nine interactions involved proteins implicated in DNA replication. Three of these were interactions among subunits of the ribonucleotide reductase protomer, a complex composed of two virus-encoded homodimers (18, 19). We observed self-association of F4L (small subunit) and of I4L (large subunit), as well as an interaction between F4L and I4L, which was positive in the reciprocal orientations of the two-hybrid vectors. Additionally, we observed self-association of F2L (deoxyuridine triphosphatase), which was previously shown to be a trimer (20); J2R (thymidine kinase), which is a known homotetramer (21); and I3L (a single-stranded DNA-binding protein) (Fig. 2A), which is thought to be monomeric based on gel filtration and electron microscopy (22, 23).
Figure 2.
Representative two-hybrid interactions. Diploids containing an ORF from the vaccinia virus activation domain array (Leu+) and a test vaccinia ORF in the DNA-binding domain vector (Trp+) were plated onto synthetic medium lacking leucine, tryptophan, and histidine and supplemented with 3 mM 3-amino-1,2,4-triazole. Each member of the vaccinia activation domain array is represented as four independent colonies, and two-hybrid positives are observed as growth of three or four diploid colonies over a background of nonreproducible single colonies. (A) I3L self-association. (B) A20R interaction with H5R, D5R, and D4R. (C) H5R interaction with G2R and B1R. (D) A ORF D interaction with A23R. (E) D7R interaction with A32L, lower cluster (arrow) is a false positive. (F) A12L interaction with A19L.
The newly identified heterodimeric interactions within this set all involved A20R, a protein that is conserved among poxviruses. Unpublished data cited by Traktman (5) suggest that the protein acts as a processivity factor for DNA synthesis in vitro. A20R bound to D4R, a uracil DNA glycosylase (24), D5R, a DNA-dependent ATPase (25), and H5R, a viral late transcription factor (Fig. 2B) (26, 27). Interestingly, H5R had been shown to associate with A18R, a negative RNA elongation factor (26). The association of A20R with D4R and D5R proteins is consistent with data showing that lethal mutations of either of these two genes blocked viral DNA replication (28, 29) and support the possibility of a multicomponent DNA replication complex. Such protein–protein interactions could account for the surprising finding that the vaccinia virus DNA glycosylase, a repair enzyme, is essential for DNA synthesis.
Transcription.
Nine interactions involved proteins that have been implicated in transcription. Several of these associations were previously observed or predicted whereas others are novel. As also reported by yeast two-hybrid analysis (26), we found that the late transcription elongation factor G2R bound to H5R, a late transcription factor. Of special note, we found that H5R also interacted with B1R, a protein kinase (Fig. 2C) (30, 31). This interaction supports previous data suggesting that H5R is phosphorylated by B1R (32). Furthermore, A49R, a protein for which no function has been determined, interacted with H5R. This interaction suggests a potential role for A49R in late transcription. However, in view of our finding that H5R interacts with A20R, a putative DNA synthesis processivity factor, it is interesting that the A49R ORF is interposed between the genes for thymidylate kinase and DNA ligase.
As anticipated, the subunits A23R and A8R of the heterodimeric intermediate transcription factor VITF-3 (33) were found to interact in the yeast two-hybrid system. Unexpectedly, however, a small ORF of 306 bp called A ORF D (3) also bound to A23R (Fig. 2D). Interestingly, A ORF D is situated on the opposite strand of the A7L ORF, which encodes a subunit of the early transcription factor VETF and would therefore not be expected to be expressed. The interaction of A1L and G8R, two of the three late transcription factors (34), is reasonable although a physical association had not been demonstrated in any previous studies. G8R was also found to self-associate. We also observed the binding of D7R, an RNA polymerase subunit (35, 36), to A32L, a protein that has been associated with virion morphogenesis and DNA packaging (37) (Fig. 2E). Unexpectedly, we identified an interaction of L4R with F8L, observed in the reciprocal orientations of the two-hybrid vectors. L4R is a major core component of virus particles (7) that binds single-stranded DNA as well as RNA and stimulates the DNA helicase activity of I8R (38, 39). When expression of L4R was repressed, noninfectious virus particles defective in transcription were produced (40, 41). It was suggested that L4R may be involved in unwinding the DNA template for transcription. F8L has a limited region of homology with the proline repeat region of iActA, a Listeria ivannovii protein involved in actin tail formation, although no effect on actin tail formation was observed in F8L deletion strains (42). A functional relationship between L4R and F8L remains to be determined.
Virion Structural/Morphogenesis Proteins.
Vaccinia virus particles comprise membrane and non-membrane proteins. Many of the latter are contained within a complex core structure and include enzymes and factors involved in mRNA synthesis. The roles of abundant proteins are assumed to be structural although they may have additional roles. The precursors of some major core proteins are proteolytically processed during the assembly of vaccinia virus particles (43). One of these, P4A, encoded by the A10L ORF (44, 45), can interact with itself in the two-hybrid system. The A12L protein is a virion component that also undergoes proteolytic processing as determined by N-terminal sequencing analysis (7). It was found to interact with A19L (Fig. 2F), a protein of unknown function. The putative DNA-packaging protein A32L required for virion morphogenesis (37) was found to associate with A11R, a protein of unknown function, in addition to the RNA polymerase subunit D7R. A4L, a virion protein that localizes between the core and membrane (46), associated with A45R, a superoxidase dismutase homolog with no catalytic potential (47, 48). In addition, A45R was found to self-associate, fulfilling a prediction of J. Tainer (personal communication) based on the fitting of its amino acid sequences with the known dimeric structure of superoxide dismutase (49). J1R, another virion protein (50), also interacted in the yeast two-hybrid system with the superoxide dismutase homolog. Because J1R exhibited self-interaction, homodimers of J1R may interact with homodimers of A45R. Although the superoxide dismutase homolog has not been localized, based on its associations, we predict that it will be a virion protein.
The approach used here, of expressing entire ORFs including the hydrophobic segments, probably contributed to the difficulty in detecting interactions of membrane proteins with this nuclear activation system. Only one bona fide membrane protein, the product of the A14L ORF, was identified. This protein, which is required for formation of virus envelopes (51), interacted with the uncharacterized A40R protein, which is a natural killer receptor homolog and contains a putative lectin domain (50).
Virus–Host Interactions.
Poxviruses encode a large number of proteins that are used to counter host defense mechanisms. Many of these viral proteins interact with cell proteins, perhaps accounting in part for the small number of interactions with other viral proteins found in this study. We detected the self-association of E3L, a double-stranded RNA-binding protein involved in IFN resistance (52). This interaction had been previously described, as well as that between E3L and the substrate binding domain of the host IFN-induced double-stranded RNA-activated protein kinase (53, 54). An interaction between the K1L protein, required for replication in rabbit kidney and human cell lines (55), with the uncharacterized C10L protein was recently found by using another version of the yeast two-hybrid system (A. Grunhaus and B.M., unpublished data) and was confirmed in the present screening. The biological relationship between the K1L and C1OL proteins remains to be determined. The A39R protein is a member of the semaphorin family and was used to isolate a semaphorin receptor from a human B cell line (56). A soluble form of A39R up-regulated intercellular adhesion molecule-1 and induced cytokine production from human monocytes (56). We identified an interaction between the A39R protein and E7R, a myristylated protein of unknown function (57) with no discernible homology to the semaphorin receptor. The profilin homolog A42R binds with high affinity to polyphosphoinositides, suggesting that it has a role in regulating the metabolism of these signaling molecules in vivo (47, 58). We found that A42R interacted with A26L, a protein related to the cowpox A type inclusion protein involved in sequestering intracellular virions (59, 60).
Conclusions
We have reported all interactions of full-length vaccinia virus proteins with each other that could be detected by this version of the yeast two-hybrid assay. For many reasons, the 37 interactions found represent only a fraction of those occurring during a viral infection. Modifications of this approach, e.g., by removing hydrophobic domains or by using other interaction systems that do not rely on nuclear transport, would increase this number. As nine of the interactions found had been previously observed, it seems likely that many of the others are also biologically relevant. Although it will be essential to confirm and extend our findings by other means, we believe that these data provide a launching point for the analysis of a large number of poxvirus proteins for which no function is yet known.
Acknowledgments
We thank Stephanie A. Monks for her assistance in the statistical analysis of the data, John Robinson for excellent technical support, and George Katsafanas for preparing the vaccinia virus DNA. We also thank Tatiana Senkevich and Mark Challberg for comments on the manuscript. This work was supported by a grant from the Merck Genome Research Institute and a grant from the National Institutes of Health (GM54415). S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080078197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080078197
References
- 1.Moss B. In: Fields Virology. Fields N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 2.Moss B. Proc Natl Acad Sci USA. 1996;93:11341–11348. doi: 10.1073/pnas.93.21.11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. , 517–563. [DOI] [PubMed] [Google Scholar]
- 4.Moss B. In: Transcription Mechanisms and Regulation. Conaway R, Conaway J, editors. New York: Raven; 1993. pp. 185–205. [Google Scholar]
- 5.Traktman P. In: DNA Replication in Eukaryotic Cells. DePamphilis M L, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 775–793. [Google Scholar]
- 6.Jensen O N, Houthaeve T, Shevchenko A, Cudmore S, Ashford T, Mann M, Griffiths G, Krijnse Locker J. J Virol. 1996;70:7485–7497. doi: 10.1128/jvi.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi T, Oie M, Ichihashi Y. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- 8.Alcami A, Symons J A, Khanna A, Smith G L. Semin Virol. 1998;8:419–427. [Google Scholar]
- 9.McFadden G, Barry M. Semin Virol. 1998;8:429–442. [Google Scholar]
- 10.Fenner F. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott-Raven; 1996. pp. 2673–2702. [Google Scholar]
- 11.Hudson J R, Jr, Dawson E P, Rushing K L, Jackson C H, Lockshon D, Conover D, Lanciault C, Harris J R, Simmons S J, Rothstein R, Fields S. Genome Res. 1997;7:1169–1173. doi: 10.1101/gr.7.12.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel P L, Roecklein J A, SenGupta D, Fields S. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 13.Earl P L, Moss B, Wyatt L S, Carroll M W. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Greene & Wiley Interscience; 1998. pp. 16.17.1–16.17.19. [Google Scholar]
- 14.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman F. Methods Enzymol. 1991;194:3–20. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 16.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivansan M, Pochart P, et al. Nature (London) 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 17.Ma H, Kunes S, Schatz P J, Botstein D. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 18.Howell M L, Sanders-Loehr J, Loehr T M, Roseman N A, Mathews C K, Slabaugh M B. J Biol Chem. 1992;267:1705–1711. [PubMed] [Google Scholar]
- 19.Slabaugh M B, Davis R E, Roseman N A, Mathews C K. J Biol Chem. 1993;268:17803–17810. [PubMed] [Google Scholar]
- 20.Roseman N A, Evans R K, Mayer E L, Rossi M A, Slabaugh M B. J Biol Chem. 1996;271:23506–23511. doi: 10.1074/jbc.271.38.23506. [DOI] [PubMed] [Google Scholar]
- 21.Black M E, Hruby D E. Biochem Biophys Res Commun. 1990;169:1080–1086. doi: 10.1016/0006-291X(90)92005-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rochester S C, Traktman P. J Virol. 1998;72:2917–2926. doi: 10.1128/jvi.72.4.2917-2926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng M, Palaniyar N, Zhang W, Evans D H. J Biol Chem. 1999;274:21637–21644. doi: 10.1074/jbc.274.31.21637. [DOI] [PubMed] [Google Scholar]
- 24.Upton C, Stuart D T, McFadden G. Proc Natl Acad Sci USA. 1993;90:4518–4522. doi: 10.1073/pnas.90.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans E, Klemperer N, Ghosh R, Traktman P. J Virol. 1995;69:5353–5361. doi: 10.1128/jvi.69.9.5353-5361.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black E P, Moussatche N, Condit R C. Virology. 1998;245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- 27.Kovacs G R, Moss B. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellison K S, Peng W, McFadden G. J Virol. 1996;70:7965–7973. doi: 10.1128/jvi.70.11.7965-7973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans E, Traktman P. Chromosoma. 1992;102:S72–S82. doi: 10.1007/BF02451789. [DOI] [PubMed] [Google Scholar]
- 30.Lin S, Chen W, Broyles S S. J Virol. 1992;66:2717–2723. doi: 10.1128/jvi.66.5.2717-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rempel R E, Traktman P. J Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaud G, Beaud R, Leader D P. J Virol. 1995;69:1819–1826. doi: 10.1128/jvi.69.3.1819-1826.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanz P, Moss B. Proc Natl Acad Sci USA. 1999;96:2692–2697. doi: 10.1073/pnas.96.6.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keck J G, Baldick C J, Jr, Moss B. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- 35.Ahn B Y, Jones E V, Moss B. J Virol. 1990;64:3019–3024. doi: 10.1128/jvi.64.6.3019-3024.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quick S D, Broyles S S. Virology. 1990;178:603–605. doi: 10.1016/0042-6822(90)90362-u. [DOI] [PubMed] [Google Scholar]
- 37.Cassetti M C, Merchlinsky M, Wolffe E J, Weisberg A S, Moss B. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayliss C D, Smith G L. Nucleic Acids Res. 1997;25:3984–3990. doi: 10.1093/nar/25.20.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bayliss C D, Wilcock D, Smith G L. J Gen Virol. 1996;77:2827–2831. doi: 10.1099/0022-1317-77-11-2827. [DOI] [PubMed] [Google Scholar]
- 40.Wilcock D, Smith G L. J Virol. 1996;70:934–943. doi: 10.1128/jvi.70.2.934-943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilcock D, Smith G L. Virology. 1994;202:294–304. doi: 10.1006/viro.1994.1346. [DOI] [PubMed] [Google Scholar]
- 42.Higley S, Way M. J Gen Virol. 1997;78:2633–2637. doi: 10.1099/0022-1317-78-10-2633. [DOI] [PubMed] [Google Scholar]
- 43.Moss B, Rosenblum E N. J Mol Biol. 1973;81:267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- 44.Van Meir E, Wittek R. Arch Virol. 1988;102:19–27. doi: 10.1007/BF01315559. [DOI] [PubMed] [Google Scholar]
- 45.Vanslyke J K, Whitehead S S, Wilson E M, Hruby D E. Virology. 1991;183:467–478. doi: 10.1016/0042-6822(91)90976-i. [DOI] [PubMed] [Google Scholar]
- 46.Cudmore S, Blasco R, Vincentelli R, Esteban M, Sodeik B, Griffiths G, Krijnse Locker J. J Virol. 1996;70:6909–6921. doi: 10.1128/jvi.70.10.6909-6921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blasco R, Cole N B, Moss B. J Virol. 1991;65:4598–4608. doi: 10.1128/jvi.65.9.4598-4608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith G L, Chan Y S, Howard S T. J Gen Virol. 1991;72:1349–1376. doi: 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 49.Parge H E, Hallewell R A, Tainer J A. Proc Natl Acad Sci USA. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antoine G, Scheiflinger F, Dorner F, Falkner F G. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez J R, Risco C, Carrascosa J L, Esteban M, Rodriguez D. J Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang H W, Watson J C, Jacobs B L. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho C K, Shuman S. Virology. 1996;217:272–284. doi: 10.1006/viro.1996.0114. [DOI] [PubMed] [Google Scholar]
- 54.Sharp T V, Moonan F, Romashko A, Joshi B, Barber G N, Jagus R. Virology. 1998;250:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 55.Perkus M E, Goebel S J, Davis S W, Johnson G P, Limbach K, Norton E K, Paoletti E. Virology. 1990;179:276–286. doi: 10.1016/0042-6822(90)90296-4. [DOI] [PubMed] [Google Scholar]
- 56.Comeau M R, Johnson R, DuBose R F, Petersen M, Gearing P, VandenBos T, Park L, Farrah T, Buller R M, Cohen J I, et al. Immunity. 1998;8:473–482. doi: 10.1016/s1074-7613(00)80552-x. [DOI] [PubMed] [Google Scholar]
- 57.Martin K H, Grosenbach D W, Franke C A, Hruby D E. J Virol. 1997;71:5218–5226. doi: 10.1128/jvi.71.7.5218-5226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Machesky L M, Cole N B, Moss B, Pollard T D. Biochemistry. 1994;33:10815–10824. doi: 10.1021/bi00201a032. [DOI] [PubMed] [Google Scholar]
- 59.Amegadzie B Y, Sisler J R, Moss B. Virology. 1992;186:777–782. doi: 10.1016/0042-6822(92)90046-r. [DOI] [PubMed] [Google Scholar]
- 60.Funahashi S, Sato T, Shida H. J Gen Virol. 1988;69:35–47. doi: 10.1099/0022-1317-69-1-35. [DOI] [PubMed] [Google Scholar]