Abstract
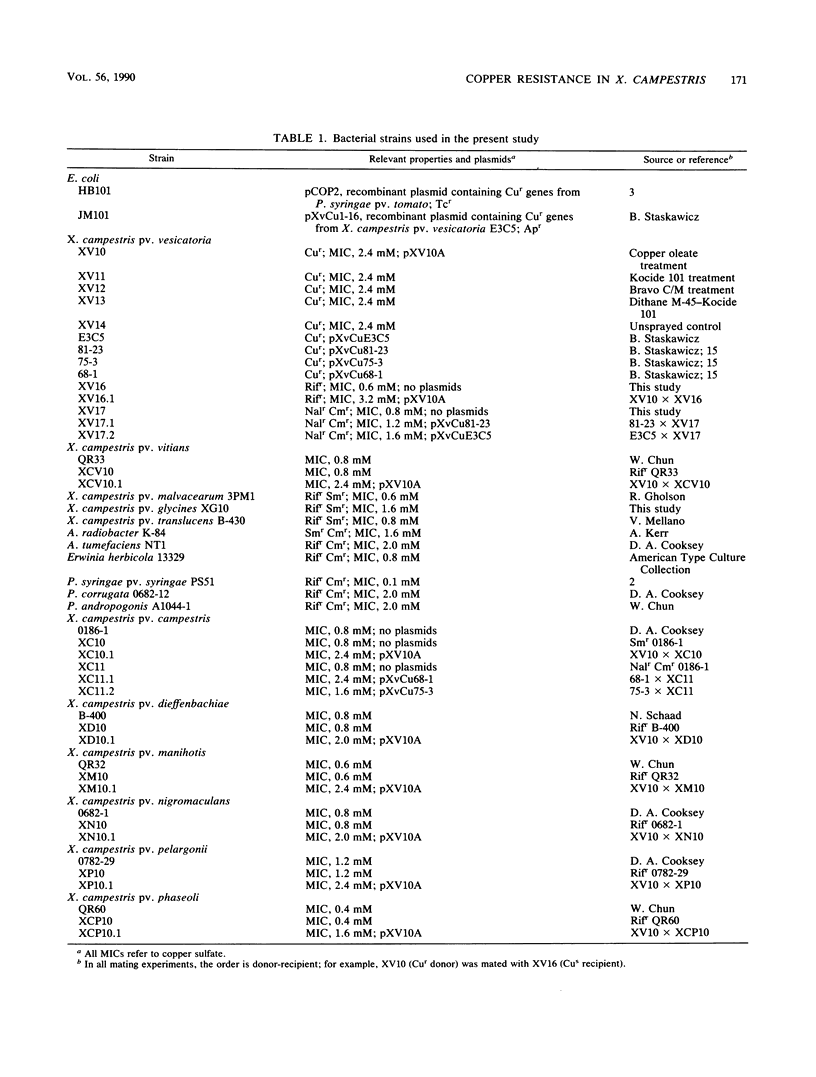
The efficacy of copper bactericides for control of Xanthomonas campestris pv. vesicatoria in eastern Oklahoma tomato fields was evaluated. Copper bactericides did not provide adequate control, and copper-resistant (Cur) strains of the pathogen were isolated. The Cur genes in these strains were located on a large indigenous plasmid designated pXV10A. The host range of pXV10A was investigated; this plasmid was efficiently transferred into 8 of 11 X. campestris pathovars. However, the transfer of pXV10A to other phytopathogenic genera was not detected. DNA hybridization experiments were performed to characterize the Cur genes on pXV10A. A probe containing subcloned Cur genes from X. campestris pv. vesicatoria E3C5 hybridized to pXV10A; however, a subclone containing Cur genes from P. syringae pv. tomato PT23 failed to hybridize to pXV10A. Further DNA hybridization experiments were performed to compare pXV10A with pXvCu plasmids, a heterogenous group of Cur plasmids present in strains of X. campestris pv. vesicatoria from Florida. These studies indicated that the Cur genes on pXV10A and pXvCu plasmids share nucleotide sequence homology and may have a common origin. Further experiments showed that these plasmids are distinctly different because pXV10A did not contain sequences homologous to IS476, an insertion sequence present on pXvCu plasmids.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender C. L., Cooksey D. A. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986 Feb;165(2):534–541. doi: 10.1128/jb.165.2.534-541.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender C. L., Cooksey D. A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987 Feb;169(2):470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A. Characterization of a Copper Resistance Plasmid Conserved in Copper-Resistant Strains of Pseudomonas syringae pv. tomato. Appl Environ Microbiol. 1987 Feb;53(2):454–456. doi: 10.1128/aem.53.2.454-456.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erardi F. X., Failla M. L., Falkinham J. O., 3rd Plasmid-encoded copper resistance and precipitation by Mycobacterium scrofulaceum. Appl Environ Microbiol. 1987 Aug;53(8):1951–1954. doi: 10.1128/aem.53.8.1951-1954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara M., Kamio Y., Terawaki Y. Cupric ion resistance as a new genetic marker of a temperature sensitive R plasmid, Rtsl in Escherichia coli. Biochem Biophys Res Commun. 1978 May 15;82(1):74–80. doi: 10.1016/0006-291x(78)90578-8. [DOI] [PubMed] [Google Scholar]
- Rouch D., Camakaris J., Lee B. T., Luke R. K. Inducible plasmid-mediated copper resistance in Escherichia coli. J Gen Microbiol. 1985 Apr;131(4):939–943. doi: 10.1099/00221287-131-4-939. [DOI] [PubMed] [Google Scholar]
- Tetaz T. J., Luke R. K. Plasmid-controlled resistance to copper in Escherichia coli. J Bacteriol. 1983 Jun;154(3):1263–1268. doi: 10.1128/jb.154.3.1263-1268.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]