Abstract
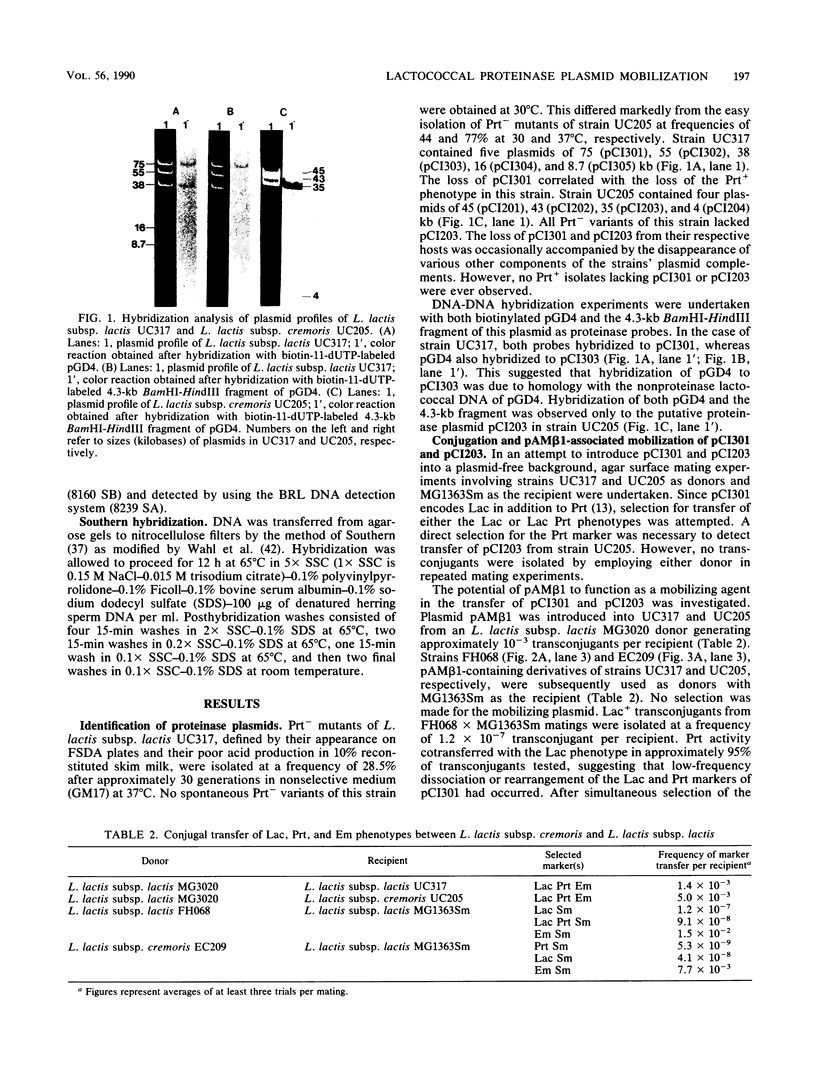
A combination of plasmid curing and DNA-DNA hybridization data facilitated the identification of proteinase plasmids of 75 (pCI301) and 35 kilobases (pCI203) in the multi-plasmid-containing strains Lactococcus lactis subsp. lactis UC317 and L. lactis subsp. cremoris UC205, respectively. Both plasmids were transferred by conjugation to a plasmid-free background only after introduction of the conjugative streptococcal plasmid, pAMβ1. All Prt+ transconjugants from matings involving either donor contained enlarged recombinant Prt plasmids. UC317-derived transconjugants were separable into different classes based on the presence of differently sized cointegrate plasmids and on segregation of the pCI301-derived Lac and Prt markers. All UC205-derived transconjugants harbored a single enlarged plasmid that was a cointegrate between pCI203 and pAMβ1. The identification of prt genes on pCI301 and pCI203 derivatives was achieved by a combination of restriction enzyme and hybridization analyses.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3. J Bacteriol. 1984 Jun;158(3):954–962. doi: 10.1128/jb.158.3.954-962.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald G. F., Gasson M. J. In vivo gene transfer systems and transposons. Biochimie. 1988 Apr;70(4):489–502. doi: 10.1016/0300-9084(88)90085-5. [DOI] [PubMed] [Google Scholar]
- Gasson M. J., Davies F. L. High-frequency conjugation associated with Streptococcus lactis donor cell aggregation. J Bacteriol. 1980 Sep;143(3):1260–1264. doi: 10.1128/jb.143.3.1260-1264.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Genetic transfer systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1983 Sep;49(3):275–282. doi: 10.1007/BF00399503. [DOI] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Daly C., Fitzgerald G. F. Identification of the Minimal Replicon of Lactococcus lactis subsp. lactis UC317 Plasmid pCI305. Appl Environ Microbiol. 1990 Jan;56(1):202–209. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M. Characterization of a pAM beta 1 deletion derivative isolated from Lactobacillus casei after conjugation. Biochimie. 1988 Apr;70(4):553–558. doi: 10.1016/0300-9084(88)90092-2. [DOI] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J., Venema G. Genetics of proteinases of lactic acid bacteria. Biochimie. 1988 Apr;70(4):475–488. doi: 10.1016/0300-9084(88)90084-3. [DOI] [PubMed] [Google Scholar]
- Kok J., van Dijl J. M., van der Vossen J. M., Venema G. Cloning and expression of a Streptococcus cremoris proteinase in Bacillus subtilis and Streptococcus lactis. Appl Environ Microbiol. 1985 Jul;50(1):94–101. doi: 10.1128/aem.50.1.94-101.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc D. J., Lee L. N. Physical and genetic analyses of streptococcal plasmid pAM beta 1 and cloning of its replication region. J Bacteriol. 1984 Feb;157(2):445–453. doi: 10.1128/jb.157.2.445-453.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lereclus D., Menou G., Lecadet M. M. Isolation of a DNA sequence related to several plasmids from Bacillus thuringiensis after a mating involving the Streptococcus faecalis plasmid pAM beta 1. Mol Gen Genet. 1983;191(2):307–313. doi: 10.1007/BF00334831. [DOI] [PubMed] [Google Scholar]
- Lereclus D., Ribier J., Klier A., Menou G., Lecadet M. M. A transposon-like structure related to the delta-endotoxin gene of Bacillus thuringiensis. EMBO J. 1984 Nov;3(11):2561–2567. doi: 10.1002/j.1460-2075.1984.tb02174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Keeler C. L., Jr, Jones K. R., Wood P. H. Molecular characterization of unique deletion mutants of the streptococcal plasmid, pAM beta 1. Plasmid. 1980 Jul;4(1):8–16. doi: 10.1016/0147-619x(80)90079-7. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Efstathiou J. D. Transductional evidence for plasmid linkage of lactose metabolism in streptococcus lactis C2. Appl Environ Microbiol. 1976 Jul;32(1):45–52. doi: 10.1128/aem.32.1.45-52.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Simultaneous loss of proteinase- and lactose-utilizing enzyme activities in Streptococcus lactis and reversal of loss by transduction. Appl Microbiol. 1974 Sep;28(3):342–346. doi: 10.1128/am.28.3.342-346.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L. Functional properties of plasmids in lactic streptococci. Antonie Van Leeuwenhoek. 1983 Sep;49(3):259–274. doi: 10.1007/BF00399502. [DOI] [PubMed] [Google Scholar]
- Novel M., Huang D. C., Novel G. Transposition of the Streptococcus lactis ssp. lactis Z270 lactose plasmid to pVA797: demonstration of an insertion sequence and its relationship to an inverted repeat sequence isolated by self-annealing. Biochimie. 1988 Apr;70(4):543–551. doi: 10.1016/0300-9084(88)90091-0. [DOI] [PubMed] [Google Scholar]
- Polzin K. M., Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987 Dec;169(12):5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Slos P., Robert C., Castellino I., Mercenier A. Conjugative mobilization as an alternative vector delivery system for lactic streptococci. Appl Environ Microbiol. 1987 Oct;53(10):2405–2413. doi: 10.1128/aem.53.10.2405-2413.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. D., Clewell D. B. Return of Streptococcus faecalis DNA cloned in Escherichia coli to its original host via transformation of Streptococcus sanguis followed by conjugative mobilization. J Bacteriol. 1984 Dec;160(3):1109–1114. doi: 10.1128/jb.160.3.1109-1114.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelie D., Venema G. Bacillus subtilis generates a major specific deletion in pAM beta 1. Appl Environ Microbiol. 1987 Oct;53(10):2458–2463. doi: 10.1128/aem.53.10.2458-2463.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]