Abstract
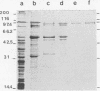
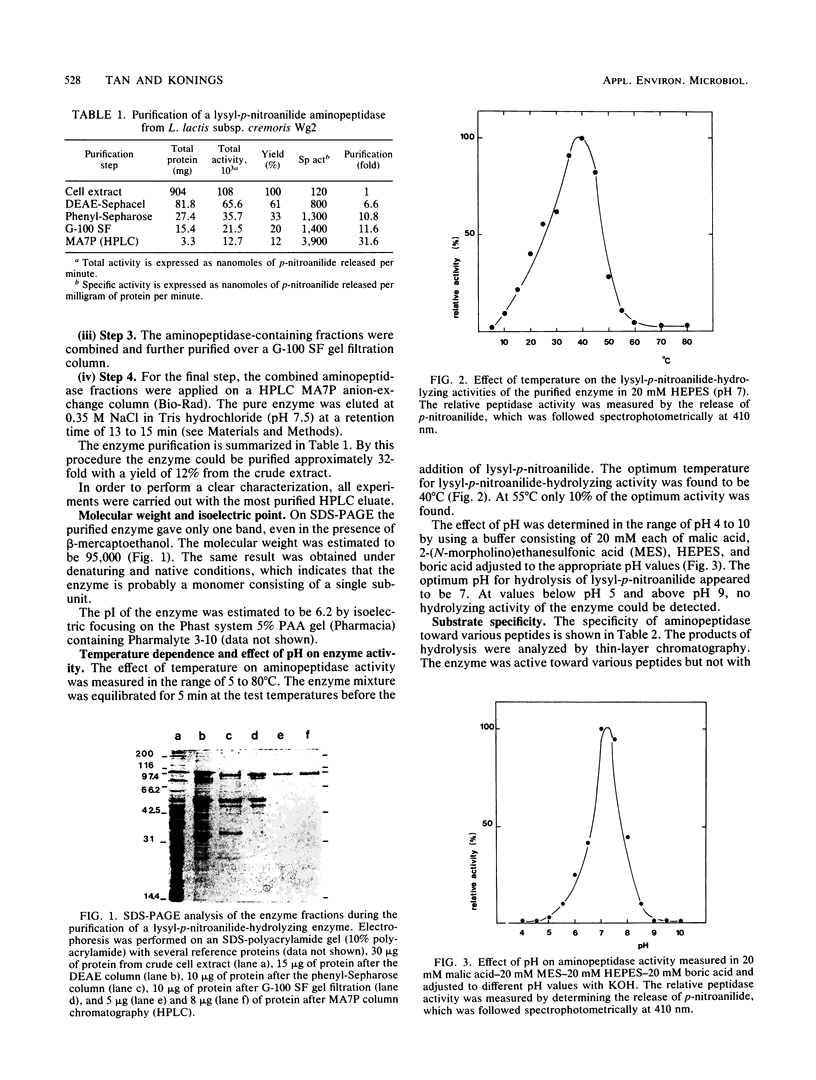
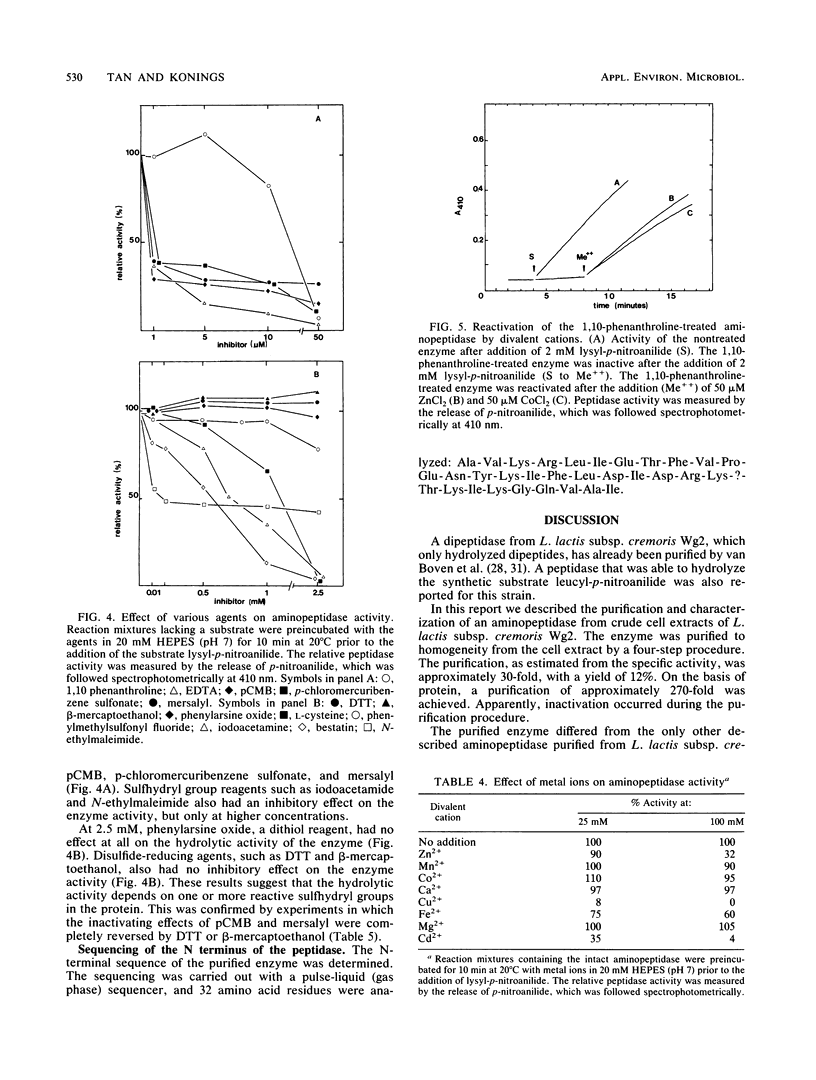
An aminopeptidase was purified to homogeneity from a crude cell extract of Lactococcus lactis subsp. cremoris Wg2 by a procedure that included diethyl-aminoethane-Sephacel chromatography, phenyl-Sepharose chromatography, gel filtration, and high-performance liquid chromatography over an anion-exchange column. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the purified enzyme showed a single protein band with a molecular weight of 95,000. The aminopeptidase was capable of degrading several peptides by hydrolysis of the N-terminal amino acid. The peptidase had no endopeptidase or carboxypeptidase activity. The aminopeptidase activity was optimal at pH 7 and 40°C. The enzyme was completely inactivated by the p-chloromecuribenzoate mersalyl, chelating agents, and the divalent cations Cu2+ and Cd2+. The activity that was lost by treatment with the sulfhydryl-blocking reagents was restored with dithiothreitol or β-mercapto-ethanol, while Zn2+ or Co2+ restored the activity of the 1,10-phenantroline-treated enzyme. Kinetic studies indicated that the enzyme has a relatively low affinity for lysyl-p-nitroanilide (Km, 0.55 mM) but that it can hydrolyze this substrate at a high rate (Vmax, 30 μmol/min per mg of protein).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Duez C., Piron-Fraipont C., Joris B., Dusart J., Urdea M. S., Martial J. A., Frère J. M., Ghuysen J. M. Primary structure of the Streptomyces R61 extracellular DD-peptidase. 1. Cloning into Streptomyces lividans and nucleotide sequence of the gene. Eur J Biochem. 1987 Feb 2;162(3):509–518. doi: 10.1111/j.1432-1033.1987.tb10669.x. [DOI] [PubMed] [Google Scholar]
- Exterkate F. A. Location of Peptidases Outside and Inside the Membrane of Streptococcus cremoris. Appl Environ Microbiol. 1984 Jan;47(1):177–183. doi: 10.1128/aem.47.1.177-183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Partial Isolation and Degradation of Caseins by Cell Wall Proteinase(s) of Streptococcus cremoris HP. Appl Environ Microbiol. 1985 Feb;49(2):328–332. doi: 10.1128/aem.49.2.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Purification and Some Properties of a Membrane-Bound Aminopeptidase A from Streptococcus cremoris. Appl Environ Microbiol. 1987 Mar;53(3):577–583. doi: 10.1128/aem.53.3.577-583.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz J., van Sinderen D., Kok J., Konings W. N. Cell Wall-Associated Proteases of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1987 Apr;53(4):853–859. doi: 10.1128/aem.53.4.853-859.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Jacques P., Frère J. M., Ghuysen J. M., Van Beeumen J. Primary structure of the Streptomyces R61 extracellular DD-peptidase. 2. Amino acid sequence data. Eur J Biochem. 1987 Feb 2;162(3):519–524. doi: 10.1111/j.1432-1033.1987.tb10670.x. [DOI] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laan H., Konings W. N. Mechanism of Proteinase Release from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1989 Dec;55(12):3101–3106. doi: 10.1128/aem.55.12.3101-3106.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Law B. A., Kolstad J. Proteolytic systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1983 Sep;49(3):225–245. doi: 10.1007/BF00399500. [DOI] [PubMed] [Google Scholar]
- Law B. A. Peptide utilization by group N streptococci. J Gen Microbiol. 1978 Mar;105(1):113–118. doi: 10.1099/00221287-105-1-113. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Monnet V., Le Bars D., Gripon J. C. Purification and characterization of a cell wall proteinase from Streptococcus lactis NCDO 763. J Dairy Res. 1987 May;54(2):247–255. doi: 10.1017/s0022029900025383. [DOI] [PubMed] [Google Scholar]
- Smid E. J., Driessen A. J., Konings W. N. Mechanism and energetics of dipeptide transport in membrane vesicles of Lactococcus lactis. J Bacteriol. 1989 Jan;171(1):292–298. doi: 10.1128/jb.171.1.292-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S., Exterkate F. A., Slangen C. J., de Veer G. J. Comparative Study of Action of Cell Wall Proteinases from Various Strains of Streptococcus cremoris on Bovine alpha(s1)-, beta-, and kappa-Casein. Appl Environ Microbiol. 1986 Nov;52(5):1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Yan T. R., Azuma N., Kaminogawa S., Yamauchi K. Purification and Characterization of a Substrate-Size-Recognizing Metalloendopeptidase from Streptococcus cremoris H61. Appl Environ Microbiol. 1987 Oct;53(10):2296–2302. doi: 10.1128/aem.53.10.2296-2302.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T. R., Azuma N., Kaminogawa S., Yamauchi K. Purification and characterization of a novel metalloendopeptidase from Streptococcus cremoris H61. A metalloendopeptidase that recognizes the size of its substrate. Eur J Biochem. 1987 Mar 2;163(2):259–265. doi: 10.1111/j.1432-1033.1987.tb10796.x. [DOI] [PubMed] [Google Scholar]
- van Boven A., Konings W. N. A Phosphate-Bond-Driven Dipeptide Transport System in Streptococcus cremoris Is Regulated by the Internal pH. Appl Environ Microbiol. 1987 Dec;53(12):2897–2902. doi: 10.1128/aem.53.12.2897-2902.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boven A., Konings W. N. Energetics of Leucyl-Leucine Hydrolysis in Streptococcus cremoris Wg(2). Appl Environ Microbiol. 1986 Jan;51(1):95–100. doi: 10.1128/aem.51.1.95-100.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boven A., Konings W. N. Utilization of dipeptides by Lactococcus lactis ssp. cremoris. Biochimie. 1988 Apr;70(4):535–542. doi: 10.1016/0300-9084(88)90090-9. [DOI] [PubMed] [Google Scholar]
- van Boven A., Tan P. S. T., Konings W. N. Purification and Characterization of a Dipeptidase from Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):43–49. doi: 10.1128/aem.54.1.43-49.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]