Abstract
Two non-coding DNA classes, introns and intergenic regions, of Drosophila melanogaster exhibit contrasting evolutionary patterns. GC content is significantly higher in intergenic regions and affects their degree of nucleotide variability. Divergence is positively correlated with recombination rate in intergenic regions, but not in introns. We argue that these differences are due to different selective constraints rather than mutational or recombinational mechanisms.
Keywords: Drosophila, non-coding DNA, GC content, selection
1. Introduction
Non-coding DNA is becoming increasingly important in studies of genome evolution, with ample evidence for functional and selective constraints in introns (INs) and intergenic regions (IGs; e.g. Halligan et al. 2004; Andolfatto 2005). Although these forces are not well characterized and generally accepted models for the evolution of INs and IGs have not been formulated, it is clear that functional requirements, and thus selective constraints, may vary between these two classes of DNA. For instance, selective constraints due to the presence of pre-mRNA secondary structures exist only in INs, not in IGs (Chen & Stephan 2006). Using a multi-locus dataset from Drosophila melanogaster, we investigate here whether these differences in selective constraints lead to differential sequence patterns, or whether sequence composition and the dynamics of nucleotide substitution in IGs and INs have primarily been shaped by mutational/recombinational processes. Since we do not find convincing explanations based on these genetic mechanisms, selection appears to be the most likely cause of the observed differential sequence patterns in IGs and INs.
2. Material and methods
We analysed the X-linked loci sequenced in an African sample (10–12 lines) of D. melanogaster (Ometto et al. 2005b) for which we could obtain homologues in both D. simulans (by sequencing or BLAST) and D. yakuba (by BLAST). Their genomic positions are based on the D. melanogaster genome release 4.2 (http://flybase.org): loci overlapping with coding regions or transposable elements were discarded. This left us with 116 fragments located in INs and 94 solely in IGs. Sequences were aligned using MegAlign (DNAstar; Madison, WI), and adjusted by eye when needed. The homologous sequences of D. simulans and D. yakuba were used to polarize polymorphisms found in D. melanogaster and the substitutions between D. melanogaster and D. simulans, respectively.
We computed basic population genetics statistics, such as θ (Watterson 1975), π (Tajima 1983), divergence and Tajima's D (Tajima 1989), using the programme NeutralityTest, kindly provided by H. Li.
Additional details are provided in electronic supplementary material.
3. Results
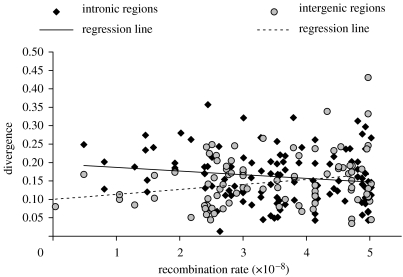
We discovered several differences in the sequence patterns of INs and IGs (summarized in tables 1–3). Although levels of polymorphism and the frequency spectrum are similar in IGs and INs (Wilcoxon test, p=0.898 and p=0.270, respectively), divergence patterns are strikingly different. IGs tend to be less diverged than INs from D. simulans (p=0.045) and D. yakuba (p=0.075). More remarkable are the discrepancies with regard to the correlation between divergence and recombination rate (Ometto et al. 2005b). When considering divergence from D. simulans, the correlation is significantly positive in IGs (Spearman's r=0.259, p=0.012), but not in INs (r=0.036, p=0.705). Due to the relatively recent split between D. melanogaster and D. simulans, such a positive correlation may simply reflect a positive correlation between polymorphism and recombination rate in their common ancestor. To test this hypothesis, we correlated divergence from D. yakuba and recombination rate (figure 1). The correlation is still present in IGs (r=0.223, p=0.031), whereas in INs it is negative (r=−0.174, p=0.062). These findings clearly show that IGs and INs experience different mutational and/or selective forces.
Table 1.
DNA variation in Drosophila non-coding DNA. Averages values per site (±1 s.e.) are reported for IGs, INs and for the combined dataset. Divergences are Jukes–Cantor corrected.
| nucleotide diversity θ | divergence from D. simulans | divergence from D. yakuba | Tajima's D | |
|---|---|---|---|---|
| INs | 0.0124±0.0006 | 0.0653±0.0025 | 0.1613±0.0065 | −0.660±0.053 |
| IGs | 0.0129±0.0008 | 0.0585±0.0030 | 0.1465±0.0073 | −0.738±0.065 |
| all | 0.0126±0.0005 | 0.0623±0.0020 | 0.1546±0.0049 | −0.695±0.041 |
Table 2.
Mutational pattern in Drosophila non-coding DNA. Average fraction (±1 s.e.) of AT→GC and GC→AT polymorphisms and substitutions.
| polymorphic in D. melanogaster | fixed in D. melanogaster | fixed in D. simulans | ||||
|---|---|---|---|---|---|---|
| AT→GC | GC→AT | AT→GC | GC→AT | AT→GC | GC→AT | |
| INs | 0.0144±0.0009 | 0.0397±0.0020 | 0.0130±0.0009 | 0.0317±0.0021 | 0.0169±0.0010 | 0.0197±0.0014 |
| IGs | 0.0155±0.0011 | 0.0394±0.0024 | 0.0147±0.0012 | 0.0252±0.0020 | 0.0157±0.0010 | 0.0164±0.0015 |
| all | 0.0149±0.0007 | 0.0396±0.0015 | 0.0137±0.0007 | 0.0288±0.0015 | 0.0164±0.0007 | 0.0182±0.0010 |
Table 3.
Summary of the main differences in the mutational pattern and nucleotide divergence between IGs and INs. See text for details.
| IGs | INs | |
|---|---|---|
| divergence from D. simulans | IGs<INs * | |
| correlation between divergence from D. simulans and recombination rate | positive * | no correlation |
| correlation between divergence from D. yakuba and recombination rate | positive * | no correlation |
| correlation between divergence along the D. melanogaster lineage and recombination rate | no correlation | no correlation |
| correlation between divergence along the D. simulans lineage and recombination rate | positive * | no correlation |
| GC content | IGs>INs * | |
| correlation between GC content and recombination rate | negative * | no correlation |
| fraction of interspecific shared alignment | IGs>INs * | |
| correlation between the fraction of interspecific shared alignment and recombination rate | negative * | no correlation |
p<0.05.
Figure 1.
Correlation between divergence from D. yakuba and recombination rate (expressed in recombination events per site per generation; Comeron et al. 1999).
To examine these differences, we analysed base composition and mutation patterns (table 2). In all three species, INs are less GC-rich than IGs (p<0.001; table 1 of electronic supplementary material). Polarizing a total of 1920 and 1564 SNPs in INs and IGs, respectively, shows that GC nucleotides exhibit a stronger tendency to mutate to AT than vice versa (p<0.0001; table 2). The analysis of the frequency spectra revealed that AT→GC polymorphisms segregate at a significantly higher average frequency (0.291±0.009;±1 s.e.) than GC→AT ones (0.256±0.006; p<0.001). This is also found when INs and IGs are considered separately (figure 1 of electronic supplementary material). However, clear differences between INs and IGs emerge when the recombination gradient is included as a variable in the analysis. In INs, recombination rate neither correlates with the frequency of AT→GC and GC→AT changes (r=−0.019, p=0.681 and r=0.054, p=0.122, respectively) nor with GC content (r=−0.086, p=0.362). The situation is distinctly different for IGs, where recombination rate shows a significantly positive correlation with the frequency of AT→GC polymorphisms (r=0.099, p=0.042; for GC→AT r=−0.001, p=0.973), and a negative one with GC content (r=−0.251, p=0.015; see also Singh et al. 2005). The first observation suggests a recombination-associated fixation bias for GC polymorphisms (e.g. GC-biased gene conversion; Galtier et al. 2006). If so, the observed negative correlation between GC content and recombination rate indicates that such bias is counteracted by other forces and/or has emerged only recently in D. melanogaster X-linked IGs. Yet, these findings do not explain the high GC content of IGs and the origin of the correlation with recombination rate.
To examine possible fixation biases, we polarized 915 and 699 fixed substitutions in D. melanogaster INs and IGs, respectively. The McDonald–Kreitman test (McDonald & Kreitman 1991) revealed that more AT→GC polymorphisms went to fixation than GC→AT ones (p=0.004 and p=0.001, for IGs and INs, respectively; two-tailed Fisher's exact test). However, we could not detect any bias in the fixation pattern to explain the difference in base composition. There is no difference in the asymmetry of the mutational pattern or in AT enrichment between IGs and INs (i.e. fixed GC→AT versus AT→GC; data not shown).
Can insertions and deletions contribute to the difference in base composition? Interestingly, fixed inserted DNA is more GC-rich than when segregating (p=0.001). In INs, it is also more GC-rich than deleted DNA (for details, see electronic supplementary material). However, when we accounted for the net gain/loss of DNA due to the action of deletions and insertions, no difference between GC enrichment in INs and IGs is evident (p>0.626 in both segregating indels or indels fixed along the D. melanogaster lineage). Thus, indel dynamics do not seem to be responsible for the contrasting pattern in base composition between INs and IGs either.
4. Discussion
The evolution of IGs and INs of Drosophila, two non-coding DNA classes, differ in subtle, but important ways (summarized in table 3). Most importantly, in the GC-rich IGs, sequence divergence tends to be lower and correlates with recombination rate, whereas the opposite is found in INs.
One explanation for the higher AT content of INs versus IGs may be a differential mutation pressure in transcribed versus non-transcribed regions, i.e. the so-called transcription-associated mutation bias (e.g. Sekelsky et al. 2000). However, we could not find differences in the mutation or fixation pattern that would support this explanation. INs do not show a higher fraction of GC→AT changes than IGs, either as polymorphisms or as fixations.
An alternative explanation for the higher AT content of INs is that we are observing ancient signatures of neutral/non-neutral forces not produced by the present mutation/substitution pattern. To investigate this possibility, we calculated GC composition at equilibrium (GC*; Sueoka 1962) inferring mutation rate from polymorphism. GC* is lower than the observed GC content (p<0.05, in INs and IGs). Thus, we cannot exclude a recent D. melanogaster-specific change in mutation bias (in line with older AT→GC polymorphisms being at higher frequency than more recent GC→AT ones; Kern & Begun 2005) or in the substitution pattern (in D. melanogaster GC→AT substitutions are more numerous than AT→GC ones, whereas the opposite is found in D. simulans, p<0.0001; see electronic supplementary material). Such changes, however, should be found in both non-coding DNAs. Since the difference in base composition between INs and IGs holds in all three species studied, they cannot explain the observations.
The contrast in substitution pattern between D. melanogaster and D. simulans and the higher frequency of AT→GC polymorphisms (relative to GC→AT ones) are also consistent with the low codon bias of D. melanogaster (favourable codons end in either G or C), which Akashi (1996) attributed to a relaxation of selection in this species. Such a relaxation, possibly due to a reduced effective population size (e.g. Ometto et al. 2005b), would have caused the fixation of otherwise non-preferred alleles (AT in this case), while the older AT→GC polymorphisms reached higher frequency. In agreement with this hypothesis, divergence calculated along the D. melanogaster lineage (see electronic supplementary material) is higher than that along the D. simulans one (p=0.0075; see also Akashi 1996; Kern & Begun 2005). This would also explain the excess of GC→AT polymorphisms relative to substitutions, with the former being more affected by a change in selection efficiency.
Next, we search for an explanation of our findings that recombination is positively correlated with nucleotide divergence (table 3). This observation may be explained by the hypothesis that recombination itself is mutagenic. However, this positive correlation holds only in IGs, and not in INs, similar to the correlation between recombination rate and θ (r=0.186, p=0.07 and r=0.049, p=0.601 for IGs and INs, respectively). Moreover, GC-rich composition appears to reduce the (speed of) accumulation of new mutations, and in IGs recombination is negatively correlated with GC. Indeed, in IGs the partial correlation coefficients (95% CI) of GC content versus divergence (controlling for recombination), GC content versus recombination (controlling for divergence), and divergence versus recombination (controlling for GC content) are −0.302 (−0.476, −0.105), −0.224 (−0.409, −0.021) and 0.172 (−0.033, 0.363), respectively. Equivalent results are obtained when θ is analysed instead of divergence (not shown). This suggests that both recombination and GC content affect the levels of nucleotide divergence and polymorphism, since each of them correlates with divergence (after controlling for the effects of the other variable). Indeed, divergence from D. simulans is negatively correlated with GC content (p<0.0001, for IGs and INs; Haddrill et al. 2005).
Is there a link between GC content and the constraints limiting divergence? Since a sequence not conserved across species is less likely to contain functionally important DNA (e.g. Ometto et al. 2005a), the absence of insertions or deletions can be used as a proxy for selective constraints. Interestingly, we found evidence that selective constraints, corresponding to blocks of less diverged DNA and possibly corresponding to functional elements, are more important in IG sequences, especially in regions with low recombination and high GC content (see electronic supplementary material). Thus, selective constraints may be involved in shaping the evolution of IGs across the recombination gradient if we assume that functional elements are primarily under purifying selection.
Recently, Andolfatto (2005) showed evidence for adaptive evolution in non-coding DNA of the X chromosome of D. melanogaster. This finding raises the intriguing possibility that the positive correlation we observed between recombination rate and divergence might be the signature of positive selection being more effective in regions of high recombination (e.g. Presgraves 2005). Divergence calculated along the D. melanogaster lineage does not correlate with recombination (r=−0.044, p=0.640 and r=0.158, p=0.128, for INs and IGs, respectively), while the one calculated along the D. simulans lineage correlates significantly (r=0.233, p=0.024 and r=0.155, p=0.096, for IGs and INs, respectively). Assuming selection is more effective in D. simulans, this result suggests that the correlation between divergence and recombination rate may be due to selective mechanisms (Akashi 1996).
Based on these arguments, it appears that different neutral and selective forces are acting on non-coding DNA (table 3). Since we could not find clear evidence for neutral (i.e. genetic) forces operating differentially in IGs versus INs, selection seems to be involved in producing the differences observed between these two regions; but, why does selection operate differently in these DNAs? The requirements for functional elements in INs and IGs are clearly different. For instance, in INs pre-mRNA secondary structures play an important role. This may lead to a form of epistatic selection with long-range fitness interactions and a relatively high AT content to avoid structures that are too stable (Chen & Stephan 2006). In contrast, in IGs directional selection at multiple, relatively independent sites (due to the modular organization of regulatory elements) may be more important, which could explain the observed correlations with recombination rate.
Acknowledgments
We are grateful to Daven Presgraves, Brian Charlesworth and two anonymous reviewers for valuable comments on an earlier version of this paper. Thanks to the Deutsche Forschungsgemeinschaft and the Volkswagen Stiftung for funding.
Supplementary Material
Additional materials and methods are explained, and we give supplementary data supporting the findings reported in the article
Polymorphism frequency spectrum in non-coding DNA (intergenic and intronic regions) of D. melanogaster
Average length and base composition across X-linked non-coding DNA fragments (intergenic and intronic regions) in Drosophila
DNA variation in Drosophila non-coding DNA (intergenic and intronic regions and the combined dataset): values are also calculated separately for the fraction of alignment shared and not shared between species
References
- Akashi H. Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster. Genetics. 1996;144:1297–1307. doi: 10.1093/genetics/144.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. doi: 10.1038/nature04107 [DOI] [PubMed] [Google Scholar]
- Chen Y, Stephan W. Weak selection on noncoding gene features. In: Fox C.W, Wolf J.B, editors. Evolutionary genetics—concepts and case studies. Oxford University Press; Oxford, UK: 2006. pp. 133–143. [Google Scholar]
- Comeron J.M, Kreitman M, Aguadé M. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Bazin E, Bierne N. GC-biased segregation of non-coding polymorphisms in Drosophila. Genetics. 2006;172:221–228. doi: 10.1534/genetics.105.046524. doi:10.1534/genetics.105.046524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill P.R, Charlesworth B, Halligan D.L, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. doi:10.1186/gb-2005-6-8-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan D.L, Eyre-Walker A, Andolfatto P, Keightley P.D. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 2004;14:273–279. doi: 10.1101/gr.1329204. doi:10.1101/gr.1329204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern A.D, Begun D.J. Patterns of polymorphism and divergence from non-coding sequences of Drosophila melanogaster and D. simulans: evidence for non-equilibrium processes. Mol. Biol. Evol. 2005;22:51–62. doi: 10.1093/molbev/msh269. doi:10.1093/molbev/msh269 [DOI] [PubMed] [Google Scholar]
- McDonald J, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. doi:10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Ometto L, Stephan W, De Lorenzo D. Insertion/deletion and nucleotide polymorphism data reveal constraints in Drosophila melanogaster introns and intergenic regions. Genetics. 2005a;169:1521–1527. doi: 10.1534/genetics.104.037689. doi:10.1534/genetics.104.037689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto L, Glinka S, De Lorenzo D, Stephan W. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 2005b;22:2119–2130. doi: 10.1093/molbev/msi207. doi:10.1093/molbev/msi207 [DOI] [PubMed] [Google Scholar]
- Presgraves D.C. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 2005;15:1651–1656. doi: 10.1016/j.cub.2005.07.065. doi:10.1016/j.cub.2005.07.065 [DOI] [PubMed] [Google Scholar]
- Sekelsky J.J, Brodsky M.H, Burtis K.C. DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell. Biol. 2000;150:F31–F36. doi: 10.1083/jcb.150.2.f31. doi:10.1083/jcb.150.2.F31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.D, Davis J.C, Petrov D.A. Codon bias and non-coding GC content correlate negatively with recombination rate on the Drosophila X chromosome. J. Mol. Evol. 2005;61:315–324. doi: 10.1007/s00239-004-0287-1. doi:10.1007/s00239-004-0287-1 [DOI] [PubMed] [Google Scholar]
- Sueoka N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl Acad. Sci. USA. 1962;85:2653–2657. doi: 10.1073/pnas.48.4.582. doi:10.1073/pnas.85.8.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. doi:10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional materials and methods are explained, and we give supplementary data supporting the findings reported in the article
Polymorphism frequency spectrum in non-coding DNA (intergenic and intronic regions) of D. melanogaster
Average length and base composition across X-linked non-coding DNA fragments (intergenic and intronic regions) in Drosophila
DNA variation in Drosophila non-coding DNA (intergenic and intronic regions and the combined dataset): values are also calculated separately for the fraction of alignment shared and not shared between species