Abstract
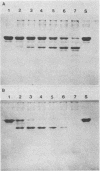
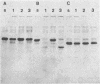
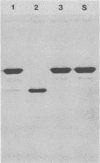
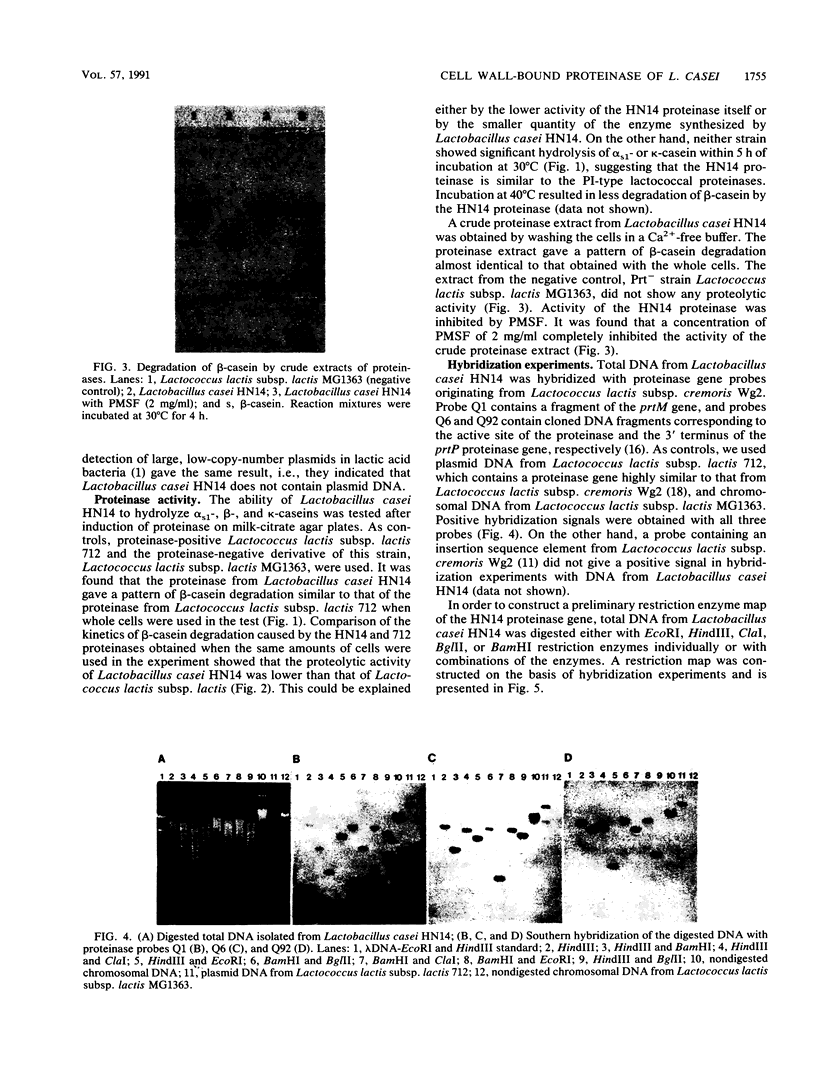
Lactobacillus casei HN14, which was isolated from homemade cheese, produces an extracellular, cell wall-bound proteinase. The HN14 proteinase can be removed from the cell envelope by washing the cells in a Ca2+-free buffer. The activity of the crude proteinase extract is inhibited by phenylmethylsulfonyl fluoride, showing that the enzyme is a serine-type proteinase. Considering the substrate specificity, the HN14 proteinase is similar to the lactococcal PI-type enzyme, since it hydrolyzes β-casein only. Lactobacillus casei HN14 appeared to be plasmid free, which suggests that the proteinase gene is chromosomally located. Chromosomal DNA of this strain hybridizes with DNA probes Q1 (which contains a fragment of the prtM gene) and Q6 and Q92 (which contain fragments of the prtP gene); all three probes originated from the proteinase gene region of Lactococcus lactis subsp. cremoris Wg2. A restriction enzyme map of the proteinase region of Lactobacillus casei HN14 was constructed on the basis of hybridization experiments. Comparison of the restriction enzyme maps of the Lactobacillus casei HN14 proteinase gene region and those of lactococcal proteinase gene regions studied so far indicates that they are highly similar.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exterkate F. A., de Veer G. J. Partial Isolation and Degradation of Caseins by Cell Wall Proteinase(s) of Streptococcus cremoris HP. Appl Environ Microbiol. 1985 Feb;49(2):328–332. doi: 10.1128/aem.49.2.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haandrikman A. J., Kok J., Laan H., Soemitro S., Ledeboer A. M., Konings W. N., Venema G. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J Bacteriol. 1989 May;171(5):2789–2794. doi: 10.1128/jb.171.5.2789-2794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haandrikman A. J., van Leeuwen C., Kok J., Vos P., de Vos W. M., Venema G. Insertion elements on lactococcal proteinase plasmids. Appl Environ Microbiol. 1990 Jun;56(6):1890–1896. doi: 10.1128/aem.56.6.1890-1896.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempler G. M., McKay L. L. Characterization of Plasmid Deoxyribonucleic Acid in Streptococcus lactis subsp. diacetylactis: Evidence for Plasmid-Linked Citrate Utilization. Appl Environ Microbiol. 1979 Feb;37(2):316–323. doi: 10.1128/aem.37.2.316-323.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):15–42. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J., Venema G. Genetics of proteinases of lactic acid bacteria. Biochimie. 1988 Apr;70(4):475–488. doi: 10.1016/0300-9084(88)90084-3. [DOI] [PubMed] [Google Scholar]
- Kok J., van Dijl J. M., van der Vossen J. M., Venema G. Cloning and expression of a Streptococcus cremoris proteinase in Bacillus subtilis and Streptococcus lactis. Appl Environ Microbiol. 1985 Jul;50(1):94–101. doi: 10.1128/aem.50.1.94-101.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan H., Konings W. N. Mechanism of Proteinase Release from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1989 Dec;55(12):3101–3106. doi: 10.1128/aem.55.12.3101-3106.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S., Exterkate F. A., Slangen C. J., de Veer G. J. Comparative Study of Action of Cell Wall Proteinases from Various Strains of Streptococcus cremoris on Bovine alpha(s1)-, beta-, and kappa-Casein. Appl Environ Microbiol. 1986 Nov;52(5):1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., van Asseldonk M., van Jeveren F., Siezen R., Simons G., de Vos W. M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989 May;171(5):2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos W. M., Vos P., de Haard H., Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989 Dec 21;85(1):169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]