Abstract
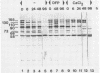
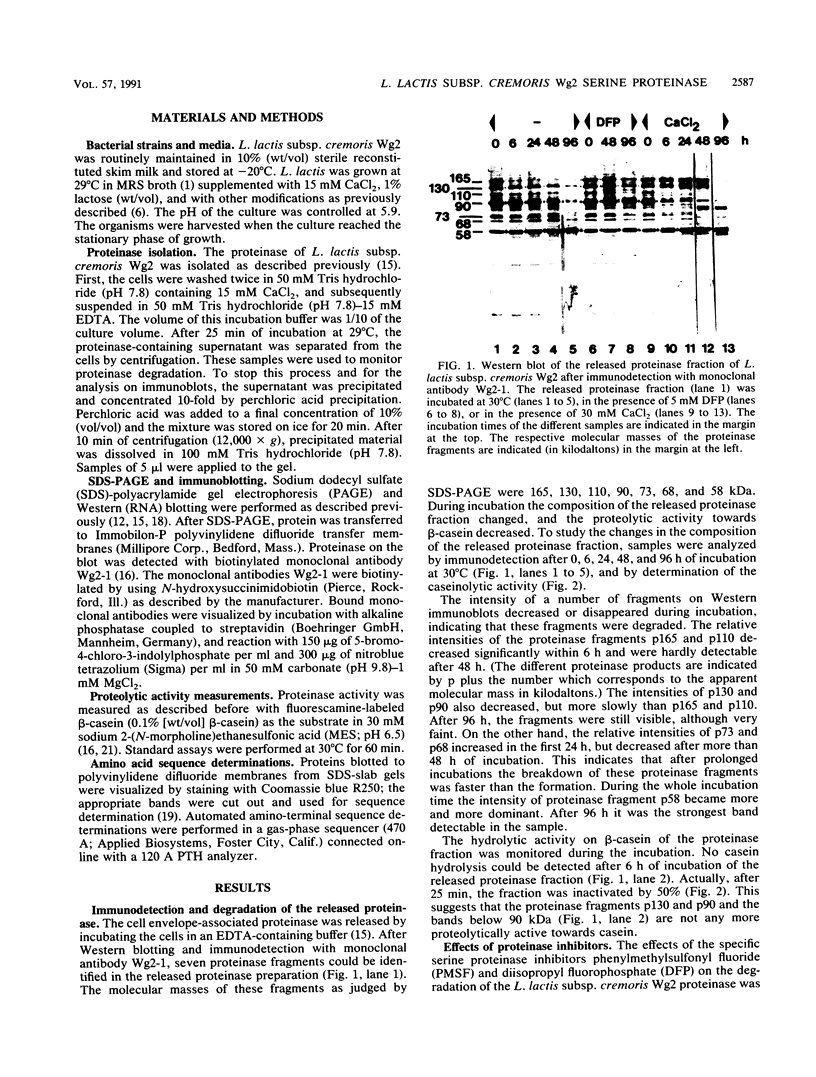
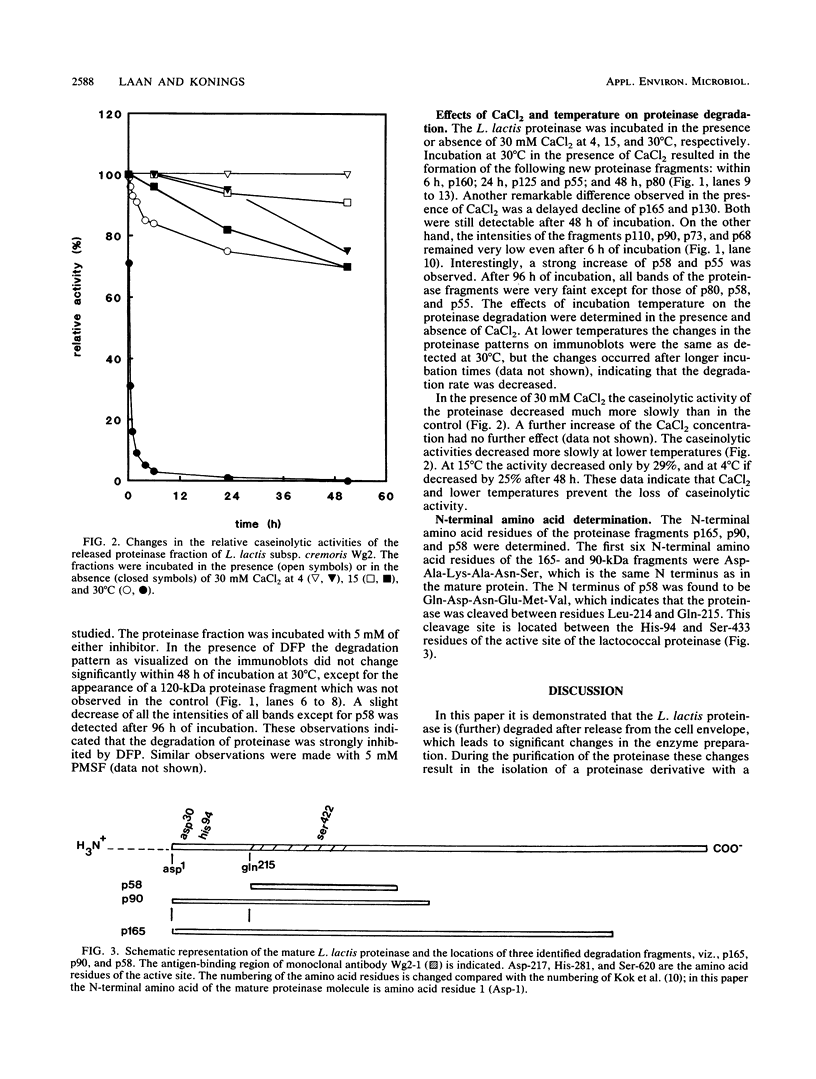
The molecular masses of purified extracellular serine proteinase of a number of Lactococcus lactis strains vary significantly, and these molecular mass values do not correspond to the values estimated on the basis of genetic data. The discrepancies can only partially be explained by N-terminal processing during maturation of the precursor enzyme and by C-terminal cleaving during the release from the cell envelope. With a monoclonal antibody that binds in the active site region of the L. lactis proteinase, the processing of the released proteinase was followed. At 30°C the proteinase was degraded with a concomitant loss of β-casein hydrolytic activity. In the presence of CaCl2, proteinase degradation was inhibited, and new degradation products were detected. The specific serine proteinase inhibitors phenylmethylsulfonyl fluoride and diisopropylfluorophosphate also inhibited proteinase degradation. Two major high-molecular-mass proteinase fragments (165 and 90 kDa) were found to have the same N-terminal amino acid sequence as the mature proteinase, i.e., [Asp-1-Ala-2-Lys-3-Ala-4-Asn-5-Ser-6, indicating that both fragments were formed by cleavage at the C terminus. The N terminus of a proteinase fragment with low molecular mass (58 kDa) started with Gln-215. In this fragment part of the active site region was eliminated, suggesting that it is proteolytically inactive. Unlike larger fragments, this 58-kDa fragment remained intact after prolonged incubations. These results indicate that autoproteolysis of the L. lactis subsp. cremoris Wg2 proteinase ultimately leads to inactivation of the proteinase by deletion of the active site region.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Haandrikman A. J., Kok J., Laan H., Soemitro S., Ledeboer A. M., Konings W. N., Venema G. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J Bacteriol. 1989 May;171(5):2789–2794. doi: 10.1128/jb.171.5.2789-2794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haandrikman A. J., Meesters R., Laan H., Konings W. N., Kok J., Venema G. Processing of the lactococcal extracellular serine proteinase. Appl Environ Microbiol. 1991 Jul;57(7):1899–1904. doi: 10.1128/aem.57.7.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz J., Exterkate F., Konings W. N. The Proteolytic Systems of Streptococcus cremoris: an Immunological Analysis. Appl Environ Microbiol. 1984 Dec;48(6):1105–1110. doi: 10.1128/aem.48.6.1105-1110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwaki M., Ikemura H., Shimizu-Kadota M., Hirashima A. Molecular characterization of a cell wall-associated proteinase gene from Streptococcus lactis NCDO763. Mol Microbiol. 1989 Mar;3(3):359–369. doi: 10.1111/j.1365-2958.1989.tb00181.x. [DOI] [PubMed] [Google Scholar]
- Kok J. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):15–42. doi: 10.1111/j.1574-6968.1990.tb04877.x. [DOI] [PubMed] [Google Scholar]
- Kok J., Hill D., Haandrikman A. J., de Reuver M. J., Laan H., Venema G. Deletion analysis of the proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):239–244. doi: 10.1128/aem.54.1.239-244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988 Jan;54(1):231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings W. N., Poolman B., Driessen A. J. Bioenergetics and solute transport in lactococci. Crit Rev Microbiol. 1989;16(6):419–476. doi: 10.3109/10408418909104474. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984 Dec;10(3-4):203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laan H., Konings W. N. Mechanism of Proteinase Release from Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1989 Dec;55(12):3101–3106. doi: 10.1128/aem.55.12.3101-3106.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan H., Smid E. J., de Leij L., Schwander E., Konings W. N. Monoclonal Antibodies to the Cell-Wall-Associated Proteinase of Lactococcus lactis subsp. cremoris Wg2. Appl Environ Microbiol. 1988 Sep;54(9):2250–2256. doi: 10.1128/aem.54.9.2250-2256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Monnet V., Le Bars D., Gripon J. C. Purification and characterization of a cell wall proteinase from Streptococcus lactis NCDO 763. J Dairy Res. 1987 May;54(2):247–255. doi: 10.1017/s0022029900025383. [DOI] [PubMed] [Google Scholar]
- Sogawa K., Takahashi K. Use of fluorescamine-labeled casein as a substrate for assay of proteinases. J Biochem. 1978 Jun;83(6):1783–1787. doi: 10.1093/oxfordjournals.jbchem.a132094. [DOI] [PubMed] [Google Scholar]
- Vos P., Simons G., Siezen R. J., de Vos W. M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989 Aug 15;264(23):13579–13585. [PubMed] [Google Scholar]
- Vos P., van Asseldonk M., van Jeveren F., Siezen R., Simons G., de Vos W. M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989 May;171(5):2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]