Abstract
Background
Polymorphism in the pfcrt gene underlies Plasmodium falciparum chloroquine resistance (CQR), as sensitive strains consistently carry lysine (K), while CQR strains carry threonine (T) at the codon 76. Previous studies have shown that microsatellite (MS) haplotype variation can be used to study the evolution of CQR polymorphism and to characterize intra- and inter-population dispersal of CQR in Papua New Guinea (PNG).
Methods
Here, following identification of new polymorphic MS in introns 2 and 3 within the pfcrt gene (msint2 and msint3, respectively), locus-by-locus and haplotype heterozygosity (H) analyses were performed to determine the distribution of this intronic polymorphism among pfcrt chloroquine-sensitive and CQR alleles.
Results
For MS flanking the pfcrt CQR allele, H ranged from 0.07 (B5M77, -18 kb) to 0.094 (9B12, +2 kb) suggesting that CQ selection pressure was responsible for strong homogenisation of this gene locus. In a survey of 206 pfcrt-SVMNT allele-containing field samples from malaria-endemic regions of PNG, H for msint2 was 0.201. This observation suggests that pfcrt msint2 exhibits a higher level of diversity than what is expected from the analyses of pfcrt flanking MS. Further analyses showed that one of the three haplotypes present in the early 1980's samples has become the predominant haplotype (frequency = 0.901) in CQR parasite populations collected after 1995 from three PNG sites, when CQR had spread throughout malaria-endemic regions of PNG. Apparent localized diversification of pfcrt haplotypes at each site was also observed among samples collected after 1995, where minor CQR-associated haplotypes were found to be unique to each site.
Conclusion
In this study, a higher level of diversity at MS loci within the pfcrt gene was observed when compared with the level of diversity at pfcrt flanking MS. While pfcrt (K76T) and its immediate flanking region indicate homogenisation in PNG CQR parasite populations, pfcrt intronic MS variation provides evidence that the locus is still evolving. Further studies are needed to determine whether these intronic MS introduce the underlying genetic mechanisms that may generate pfcrt allelic diversity.
Background
Plasmodium falciparum chloroquine resistance (CQR) was first reported in Southeast Asia and South America during the late 1950's [1]. Since then, CQR parasites have spread worldwide, with corresponding increases in malaria morbidity and mortality [2]. CQR in Papua New Guinea (PNG) was first reported in 1976 [3,4]. By the early 1980's, in vivo studies indicated that CQR P. falciparum was present in ~50% of the children, while in vitro assays revealed that ~80% of the isolates were CQR [5,6]. Recent molecular studies have shown that CQR-associated alleles had spread throughout PNG by the mid 1980's [7,8], and based on widespread chloroquine (CQ) treatment failure by the late 1990's [7,9,10], the PNG Ministry of Health changed its guidelines for malaria treatment in 2000 from CQ alone to CQ + sulphadoxine-pyrimethamine.
Genetic polymorphism associated with the CQR phenotype in P. falciparum has been identified in the P. falciparum chloroquine resistance transporter (pfcrt) gene, located on chromosome 7 [11-13]. The amino acid substitution at pfcrt codon 76 (K→T) has been shown to have the strongest association with the CQR phenotype, which can be both reversible and irreversible by verapamil [7,11,14-20]. The pfcrt gene encodes an integral membrane protein, which is localized to the parasite digestive vacuole [11] where haem molecules released during haemoglobin digestion are detoxified by the formation of haemozoin, also known as malaria pigment; CQ is suggested to interfere with this process [21,22]. P. falciparum CQR is suggested to involve mechanisms whereby pH sensitive physiologic processes inhibit formation of toxic CQ:haematin complexes in favor of haemozoin [22], or CQ efflux reduces drug concentration to the levels that are no longer parasiticidal [23-25].
In addition to pfcrt, P. falciparum multidrug resistance (pfmdr1, chromosome 5) and nine other putative transporter genes have been implicated in CQR [7,26-28]. Polymorphisms in pfmdr1 play a modulatory role in CQR [29], while those in only one of the other nine transporter genes (G7, encoding an ATP-binding cassette transporter [PlasmoDB identifier: PF13-0271]) exhibit significant association with response to the antimalarial drug artesunate [28].
To understand further the population genetics of CQR P. falciparum in PNG, samples collected from six different provinces in the early 1980's and after 1995, in both community and clinical settings, were analysed. P. falciparum samples were classified by genotyping pfcrt codons 72–76 (CVMNK represents chloroquine-sensitive [CQS] allele, SVMNT and CVIET CQR alleles), and were further analysed for pfcrt single nucleotide polymorphisms (SNPs) at codons 220 (A→S), 271 (Q→E), 326 (N→D), 356 (I→L), and 371 (R→I) [11,30], as well as 152 (T→A) and 163 (S→R), which have been recently implicated in parasite's resistance to amantadine and modification of verapamil-reversible CQR phenotype [30]. A series of four microsatellite (MS) loci and the cg2 ω repeat region flanking the pfcrt locus (-18 kb upstream to +19 kb downstream), and recently discovered MS loci within the pfcrt gene [31] were analysed to investigate the diversity and distribution of CQS and CQR alleles. MS markers occurring at regular 2–3 kb intervals in the P. falciparum genome [32] have been recently used to analyse both intra- and inter-population relationships among drug-resistant P. falciparum strains, based on the extent of linkage disequilibrium (LD) and its decay rates [33-35]. In this study, these same approaches were used to provide new insight into the dispersal and continuing evolution of CQR P. falciparum strains in PNG.
Methods
Collection of samples
Samples were collected from both placental tissues and whole blood. Seven placental tissues samples were obtained from pregnant women living in the Eastern Highlands (n = 1), East Sepik (n = 3), Manus (n = 1), Milne Bay (n = 1), and Morobe (n = 1) provinces of PNG between 1982 and 1984 [36]. Clinical data were not available for these placental samples. Peripheral blood samples were collected into K+-EDTA containing Vacutainer tubes from individuals living in three malaria-holoendemic regions of PNG [37,38]. These samples were collected from the Dreikikir region of East Sepik Province (ESP) in 1996 (n = 31), the Liksul region of Madang Province in 1996 (n = 22), and the Wosera region of ESP. The Wosera samples were collected in 1998 (n = 65) and 2002 (n = 182) during community surveys, and between 2001 and 2003 (n = 980) from symptomatic (e.g., fever, parasitaemia) patients at the local health centers. Study protocols were reviewed and approved by the Medical Research Advisory Committee, Department of Health PNG, and the University Hospitals of Cleveland Institutional Review Board.
Genomic DNA preparation and parasite reference strains
DNA was extracted from the placental tissue samples by a standard phenol/chloroform extraction method [36], and from the whole blood (200 μl) using the QIAamp 96 Blood Kit (QIAGEN, Valencia, CA). For MS allele analyses, genomic DNA preparations of six P. falciparum laboratory-adapted strains (HB3, 3D7, Dd2, K1, 7G8, and PNG1917) were used as references.
PCR amplification and genotyping of pfcrt codons
Polymerase Chain Reactions (PCR) to amplify pfcrt codons 72–76, 220, 271, 326, 356, and 371 were performed as previously described [7]. PCR to amplify the codons 152 and 163 were performed using the primers and conditions described in Additional File 1. PCR products were genotyped for the above polymorphic codons using ligase detection reaction-fluorescent microsphere assay (LDR-FMA) as recently described [39-41]. The allele-specific probes used in LDR-FMA are provided in Additional File 2.
PCR amplification and genotyping of cg2 ω repeat region and MS loci
Amplification and genotyping of MS flanking pfcrt (B5M77, 2E10, 9B12, and 2H4), cg2 ω repeat region, and a putatively neutral locus PfPK2 (chromosome 12) were performed using semi-nested PCR strategies as previously described [8,42]. Pfcrt intronic MS msint1, also known as B5M47 [13], and the newly discovered intronic MS msint2 and msint3 were amplified using nested PCR strategies; primer sequences and conditions for these amplifications are provided in Additional File 1. For each MS, one of the nest-2 amplification primers was 5' end-labeled with Cy5. PCR products were mixed 3:1 (vol/vol) with denaturing loading dye buffer (formamide 10 ml, bromophenol blue 10 mg, 0.5 M EDTA [pH 8.0] 200 μl) and denatured at 95°C for 10 min. Denatured products were run on a 6% denaturing polyacrylamide gel (6.3 M urea/32% formamide) for 3 h in a Gibco BRL sequencing apparatus (model S2, Gibco BRL Life Technologies) at 1900 V. The Cy5-labeled amplicons were visualized on the Storm 860 scanner using the software ImageQuant v5.2 (Molecular Dynamics, Sunnyvale, CA). Alleles present in the field samples were compared with those present in the six reference strains, and were numerically designated from 1 to 10, corresponding to their relative electrophoretic mobility positions on the gel (1 = slowest, largest product; 10 = fastest, smallest product). Base pair sizes of msint2 and msint3 PCR products were determined for some of the reference strains (3D7 = 222 and 200; K1 = 190 and 184; PNG1917 = 217 and 147, respectively) (Su X-Z, personal communication). However, in this study, numbers from 1 to 10 were used to designate the alleles [8]. ImageQuant was used to score multiple alleles per locus if the minor fluorescent peaks were >25% of the height of the predominant peak present at each locus. Haplotypes were constructed using the predominant allele observed at each locus.
Statistical analysis
The population genetics software Arlequin v3.0 [43] was used to compute the heterozygosity (H) at each locus (value ± SD), full-length haplotype diversity (value ± SE), and genetic differentiation (Fst), which measures variation between population groups, and LD between loci. H and Fst values measure from 0 to 1, where 0 means that there is no difference between individuals or groups, and 1 means that all individuals are unique and population groups are distinct from each other.
Results
A total of 1287 samples were analysed in this study. Seven of these were placental samples, collected from the Eastern Highlands (n = 1), East Sepik (n = 3), Manus (n = 1), Milne Bay (n = 1), and Morobe (n = 1) provinces between 1982 and 1984. From our more recent field studies, 1280 blood samples were collected from the Dreikikir (n = 31), Liksul (n = 22), and Wosera (n = 1227) regions between 1996 and 2003. The P. falciparum infection status of each sample was diagnosed by genotyping pfcrt codons 72–76 using LDR-FMA. All samples from the early 1980's, Dreikikir, and Liksul surveys were P. falciparum-infected. Of the samples collected from the Wosera, 166 samples from community surveys and 595 samples from health center surveys were P. falciparum-infected. Prevalence of genotypically CQS (CVMNK) and CQR (SVMNT and/or CVIET) P. falciparum at each sample collection site is summarized in Table 1. Of all samples (n = 1287), 8.9% were infected with CQS parasites, 47.9% with CQR parasites, and 6.9% with mixed infections, while 36.2% were not infected with P. falciparum. Interestingly, the CQR-associated pfcrt allele CVIET was observed in the Wosera community samples (0.006, n = 1/166) and clinical samples (0.013, n = 8/595) collected after 2001. Previous studies did not observe the CVIET allele in any of the PNG P. falciparum-infected samples [7,8].
Table 1.
Prevalence of pfcrt alleles (codons 72–76) in malaria-endemic regions of PNG
| Early 1980'sa | Post-1995 | ||||
| Community | Clinical | ||||
| Allele | n = 7 | Liksulb n = 22 |
Dreikikirc n = 31 |
Woserad n = 166 |
Woserae n = 595 |
| CVMNK | 0.143 | 0.045 | 0.484 | 0.241 | 0.097 |
| SVMNT | 0.857 | 0.955 | 0.516 | 0.572 | 0.791 |
| CVIET | 0 | 0 | 0 | 0.006f | 0.013 |
| CVMNK + SVMNT | 0 | 0 | 0 | 0.181 | 0.086 |
| CVMNK + CVIET | 0 | 0 | 0 | 0 | 0.002 |
| SVMNT + CVIET | 0 | 0 | 0 | 0 | 0.012 |
a, b, c Samples with mixed infections were not included.
a Samples were collected from the Eastern Highlands (n = 1), East Sepik (n = 3), Manus (n = 1), Milne Bay (n = 1), and Morobe (n = 1) provinces of PNG between 1982 and 1984 and were previously reported [8].
b, c Samples were collected in 1996 and were previously reported [7].
d Samples were collected in 1998 and 2002. The 1998 samples were previously reported [7].
e Samples were collected between 2001 and 2003 in local health centers.
f Allele found only in samples taken from the 2002 group.
Pfcrt-SVMNT allele carrying samples in both the early 1980's and post-1995 groups were further analysed for SNPs at codons 220, 271, 326, 356, and 371, as well as recently reported SNPs at codons 152 and 163 in exon-3 [30] by LDR-FMA. In this expanded SNP analysis of pfcrt-SVMNT allele, S220Q271D326L356R371 was the predominant haplotype (>0.900). No polymorphism at codons 152 and 163 was observed in any of the samples. There was some variation to the S220Q271D326L356R371 haplotype in the post-1995 samples, with 220A (0.044, n = 8/181), 271E (0.011, n = 2/176), 326N (0.006, n = 1/149), 356I (0.092, n = 18/195), and 371I (0.059, n = 11/185). Since minor alleles at one or more positions were present as mixed infections, 220_271_326_356_371 minor haplotypes could not be inferred.
While performing the amplification of exon-3 region to genotype codons 152 and 163, significant variation in the size of the PCR products was observed when run on a 2% agarose gel (Figure 1). To understand what might be contributing to this size variation, simple sequence repeats in the 3D7 chromosome 7 published sequence were located (GenBank accession number AL844506). An (AT)23 repeat was found in intron-2 (nucleotide coordinates, 309138–309183), and an A40 repeat was located in intron-3 (nucleotide coordinates, 309439–309478). Due to their intronic locations within the pfcrt gene, the MS were named msint2 and msint3 (Additional File 3). Next, to determine if one or both of these MS contributed to the size variation observed for the exon-3 containing PCR products, primers were designed to amplify these sequences using nested strategies. When these products were run on a polyacrylamide gel, it was found that both msint2 and msint3 simple sequence repeats displayed size variations.
Figure 1.
Agarose gel showing PCR products with size variability. PCR products for pfcrt codons 152–163 genotyping assay showing size variation between samples. Lanes 2, 4, 6, 8, and 10 contain PNG samples collected from the Wosera in 2002. The other lanes contain laboratory-adapted strains, lane 1-Dd2, 3-7G8, 5-PNG1917, 7-PNG1905, and 9-K1.
In the samples carrying the CQS-associated CVMNK allele, eight different msint2 alleles were observed (predominant alleles #7 [0.266] and #3 [0.241]). Eight different alleles of msint3 were also observed in these samples (predominant alleles #8 [0.395] and #6 [0.296]). In the samples carrying the CQR-associated SVMNT allele, eight msint2 alleles (predominant allele #3 [0.889]) and five msint3 alleles (predominant allele #8 [0.954]) were observed. In the samples carrying the CQR-associated CVIET allele, three msint2 alleles (predominant allele #2 [0.5]) and two msint3 alleles (predominant allele #8 [0.833]) were observed. Allele frequencies for both MS in each subset of samples are presented in Additional File 4.
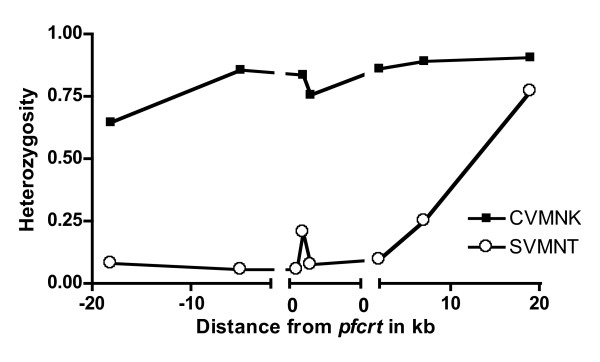
To analyse the levels of diversity at MS loci across the 40 kb region containing pfcrt-SVMNT alleles, heterozygosity (H) values were calculated for each individual locus. H ranged from 0 to 0.781 (Table 2), with the highest value observed at the 2H4 locus (0.71 to 0.781) positioned 19 kb downstream from pfcrt. Elevated H was also observed at the PfPK2 locus (0.617 to 0.769), which has been previously analysed in broad P. falciparum population surveys as a neutral marker under no apparent selection pressure [33,44]. H observed at the MS loci in the CQS (pfcrt-CVMNK) samples ranged from 0.642 to 0.903. Consistent with observations documenting reduced variation around P. falciparum genes associated with antimalarial drug resistance [45], the lowest values were observed for markers in the closest proximity to pfcrt (B5M77 to cg2; Figure 2). In contrast, elevated values were observed for the marker within pfcrt (msint2; Figure 2). Across the 40 kb region where pfcrt resides, significant LD (p < 0.05) was observed for eight of the pairwise comparisons (B5M77-B5M47, 2E10-B5M47, B5M77-pfcrt codon 371, 2E10-9B12, B5M47-9B12, B5M47-2H4, cg2ω-2H4, B5M47-PfPK2) in the CQR samples, and for four of the pairwise comparisons (pfcrt codon 220-pfcrt codon 356, 2E10-9B12, msint2-2H4, msint2-PfPK2) in the CQS samples. After a correction for multiple comparisons, significant LD was observed for two pairwise comparisons (B5M47-2H4, cg2ω-2H4; p < 0.0014) in the CQR samples, and for one pairwise comparison (pfcrt codon 220-pfcrt codon 356; p < 0.0009) in the CQS samples.
Table 2.
Heterozygosity (± standard deviation) of MS loci associated with pfcrt-SVMNTa allele in post-1995 samples.
| Locusc | Distance from pfcrt |
Liksul n = 21 |
Dreikikir n = 16 |
Woserab n = 169 |
| B5M77 | -18 kb | 0 | .342 ± .140 | .060 ± .026 |
| 2E10 | -5 kb | 0 | .154 ± .126 | .049 ± .024 |
| msint1d | 0 kb | - | - | .051 ± .034e |
| msint2 | 0 kb | .091 ± .081 | .380 ± .134 | .196 ± .039 |
| msint3 | 0 kb | 0 | .314 ± .138 | .056 ± .024 |
| 9B12 | +2 kb | .181 ± .104 | .275 ± .145 | .045 ± .030 |
| cg2 | +7 kb | .450 ± .128 | .275 ± .148 | .220 ± .042 |
| 2H4 | +19 kb | .710 ± .060 | .781 ± .102 | .746 ± .020 |
| PfPK2 | --- | .617 ± .063 | .769 ± .083 | .744 ± .019 |
a 21/22 infections in Liksul, 16/31 in Dreikikir, and 169/761 in Wosera carried the pfcrt-SVMNT allele.
b Wosera group contains both clinical and community samples.
c PCR for Liksul samples was 100% for all loci except cg2 (.905). Frequency of amplification for each locus for Dreikikir and Wosera, respectively, were B5M77, 0.875, 0.970; 2E10, 0.813, 0.941; 9B12, 0.813, 0.941; cg2, 0.813, 0.976; 2H4, 0.875, 0.935; and PfPK2, 0.813, 0.935.
d MS also known as B5M47.
e Value based only on 78 of the 82 clinical samples and no community samples.
Figure 2.
Heterozygosity of pfcrt intronic and flanking MS in pfcrt-CVMNK and SVMNT samples. Graphic representation of heterozygosity values for pfcrt intronic and flanking MS in pfcrt-CVMNK and SVMNT samples from all study sites. Data for msint1 is available only from pfcrt-SVMNT clinical samples collected from the Wosera.
Fst values were then calculated for each MS locus between the geographically distinct sample collection sites to test for population subdivision, as might be expected between locations that are separated by distance and other geographic factors that would reduce mixing of parasite populations by limiting human and/or mosquito travel. Results from these analyses (summarized in Table 3) showed similar Fst values for each pairwise comparison. Though locus-by-locus comparisons did show some significant Fst values, overall there is no clear-cut genetic variability among the three locations, suggesting no subdivision among CQR P. falciparum populations at these locations.
Table 3.
Fst values to measure genetic diversity between groups of pfcrt-SVMNT samples from post-1995 collections
| Locus | Liksul-Dreikikir | Liksul-Wosera | Dreikikir-Wosera |
| B5M77 | 0.12* | -0.01 | 0.17* |
| 2E10 | 0.04 | -0.02 | 0.04 |
| msint2 | 0.03 | -0.01 | 0.01 |
| msint3 | 0.07 | -0.01 | 0.12 |
| 9B12 | -0.04 | 0.04 | 0.10 |
| cg2 | 0.00 | 0.04 | -0.01 |
| 2H4 | 0.07* | 0.10* | 0.08* |
| PfPK2 | 0.07* | 0.01 | 0.03 |
* indicates that the value is significantly different from zero (p < 0.05).
Further, to examine the CQR parasite population structure and distribution among various locations, msint2_msint3 haplotypes in single infection samples were analysed (Table 4). In the early 1980's samples, three haplotypes were observed in the SVMNT samples (n = 6) (haplotype diversity, 0.467 ± 0.426), with the predominant haplotype being 3_8 (0.667). It was of interest to note that the other two haplotypes (8_3 and 3_2) observed in these early samples were not observed in the later samples. Among the post-1995 SVMNT samples (n = 203), 11 haplotypes were observed (haplotype diversity, 0.103 ± 0.140), and the predominant haplotype was again 3_8 (from 0.667 to 0.955). In this intronic haplotype data analysis, it was observed that while the two predominant haplotypes (3_8 and 7_8) were present at all study sites, other low frequency haplotypes were segregated and were unique to each study site (Table 4). In the CVIET samples (n = 6), three haplotypes were present (haplotype diversity, 0.533 ± 0.468), with the predominant haplotype being 2_8 (0.5). Although this intronic haplotype was also observed in association with the SVMNT allele in the Wosera community survey samples (0.011), the other two haplotypes (1_8 [0.333] and 8_7 [0.167]) were unique to the CVIET samples; the 3_8 and 7_8 haplotypes that were predominant among the SVMNT samples were not observed in the CVIET samples. Finally, the intronic MS haplotypes associated with the post-1995 CVMNK samples were examined, and a total of 27 haplotypes in 54 samples were found (haplotype diversity, 0.772 ± 0.519), showing a much higher level of diversity than that observed among the SVMNT samples (Figure 2).
Table 4.
Intronic haplotype frequencies for pfcrt-SVMNT and CVIET samplesa
| Early 1980's | Post-1995 | |||||
| Community | Clinical | |||||
| Liksul | Dreikikir | Wosera | Wosera | |||
|
Haplotype (msint2_msint3) |
SVMNT nb = 6 |
SVMNT n = 22 |
SVMNT n = 21 |
SVMNT n = 88 |
SVMNT n = 82 |
CVIET n = 6 |
| 3_8 | 0.667 | 0.955 | 0.667c | 0.898 | 0.951 | 0 |
| 7_8 | 0 | 0.045 | 0.095 | 0.057 | 0.037 | 0 |
| 8_3 | 0.167 | 0 | 0 | 0 | 0 | 0 |
| 3_2 | 0.167 | 0 | 0 | 0 | 0 | 0 |
| 7_4 | 0 | 0 | 0.048 | 0 | 0 | 0 |
| 3_3 | 0 | 0 | 0.048 | 0 | 0 | 0 |
| 3_6 | 0 | 0 | 0.048 | 0 | 0 | 0 |
| 6_6 | 0 | 0 | 0.048 | 0 | 0 | 0 |
| 10_8 | 0 | 0 | 0.048 | 0 | 0 | 0 |
| 8_8 | 0 | 0 | 0 | 0.023 | 0 | 0 |
| 9_8 | 0 | 0 | 0 | 0.011 | 0 | 0 |
| 2_8 | 0 | 0 | 0 | 0.011 | 0 | 0.500 |
| 4_8 | 0 | 0 | 0 | 0 | 0.012 | 0 |
| 1_8 | 0 | 0 | 0 | 0 | 0 | 0.333 |
| 8_7 | 0 | 0 | 0 | 0 | 0 | 0.167 |
a Total number of samples 215: early 1980's (6), Liksul (21), Dreikikir (16), Wosera community samples (85), Wosera clinical samples (81 SVMNT, 6 CVIET).
b "n" indicates the number of haplotype infections present, which may or may not be equal to the total number of samples due to mixed haplotypes in some samples. Overall, 225 haplotypes were found in 215 samples.
c Frequencies in bold indicate that the haplotype was also observed in the CQS samples from that region.
Discussion
Following the discovery of the association between pfcrt K76T and P. falciparum CQR [11], Wootton et al. evaluated a number of MS loci flanking pfcrt (± 100 kb) of 87 laboratory-adapted strains collected from malarious regions around the world to investigate the impact of CQ selection pressure on the genetic diversity in this region of the genome [35]. Results of this study comparing CQS and CQR-associated alleles showed significant reduction in MS allelic diversity, increased LD, and uniform haplotypes for the markers in the closest proximity to the pfcrt CQR allele [35]. They suggested that CQ was responsible for selection-driven "sweeps", homogenising genetic diversity at polymorphic sites in close physical linkage with CQR-associated alleles (hitchhiking) [35]. Similar observations have been made using MS flanking the alleles associated with pyrimethamine resistance at the dihydrofolate reductase gene (chromosome 4) [34,46-49] and quinine resistance at pfnhe-1, encoding a putative Na(+)/H(+) exchanger (chromosome 13) [50]. A more recent study of MS flanking pfcrt, including some of the same samples analysed here, used these approaches to describe population subdivision between the early 1980's and late 1990's CQR P. falciparum strains in PNG [8].
With these observations in mind, it was expected that very low or no heterozygosity would be observed, similar to that reported by Wootton et al. for msint1 (B5M47), at msint2 and msint3 in CQR P. falciparum strains. Interestingly, this and a previous study [31] found higher levels of MS variability within the introns of the pfcrt gene than the variability at MS loci flanking pfcrt (Figure 2; msint2 vs. 2E10 or 9B12, [0.201 vs. 0.051 or 0.094]). As the flanking and intronic MS sites are similar in their length and nucleotide content, two of the features associated with MS variability, it is not clear what genetic factors contribute to these higher levels of variability in pfcrt introns 2 and 3.
Intronic polymorphisms may provide important insights into maintenance of the pfcrt gene sequence. In addition to the MS length polymorphism reported now in pfcrt msint2, msint3 and msint4, a review of genomic sequences currently available at GenBank for P. falciparum strains 3D7 (GenBank accession number AL844506) and Dd2 (GenBank accession number AF030694) showed that simple sequence repeats were present within all pfcrt introns. A comparison between 3D7 and Dd2 genomic sequences has shown additional length polymorphism for simple sequence repeats in introns 7 (T14 vs. T13), 9 (AT18 vs. AT15), 10 (T14 vs. T15), and two in intron-11 (T18 vs. T23 and T31 vs. T19). The highly AT-rich and polymorphic characteristics of many intronic repeats has been described for a number of P. falciparum genes [51]. The high level of intronic diversity has been suggested to result from slippage of DNA polymerase [32,33]. Alternatively, repetitive AT-rich sequences that are palindromic have the potential to form intrastrand base-pairing, resulting in hairpins that are susceptible to breakage followed by meiotic recombination [52]. Events of this nature could contribute to gene conversion where polymorphic exons are swapped between parental alleles in the formation of new progeny alleles [53,54]. Further evaluation of pfcrt in laboratory adapted strains and natural P. falciparum isolates may reveal sequence-based relationships supportive of the hypothesis that these loci promote recombination/gene conversion within this genomic region [55].
In addition to evaluating pfcrt intronic MS, this study provides the first evidence of the CQR-associated CVIET allele in PNG. While CVIET is the predominant CQR-associated pfcrt allele in many parts of Southeast Asia [11,35], earlier studies in PNG [7,8] did not find this allele in any of the samples. Since the allele is found in nearby Indonesian regions [56,57], it is likely that this allele has been imported into PNG. When intronic MS loci in pfcrt-SVMNT and pfcrt-CVIET samples were compared, significant genetic differences were found between the two groups. The predominant msint2_msint3 haplotype 3_8 associated with pfcrt-SVMNT was not seen in pfcrt-CVIET samples. The haplotype diversity was significantly higher in the pfcrt-CVIET samples (0.533 ± 0.468) than in the pfcrt-SVMNT samples (0.103 ± 0.14). Further, when flanking MS haplotype (from B5M77 to 2H4) diversity was compared between the two groups of samples from the Wosera, the extended haplotype diversity was also significantly higher in the pfcrt-CVIET samples (0.7 ± 0.728) than in the pfcrt-SVMNT samples (0.06 ± 0.077). These analyses rule out the possibility that the CVIET allele in PNG has arisen on the genetic background of more prevalent SVMNT allele.
Analysis of the diversity at intronic MS provides an "inside look" at the genetic background of pfcrt from an evolutionary perspective. Among all pfcrt-SVMNT samples, the predominant msint2 allele #3 and msint3 allele #8 (msint2_msint3 haplotype 3_8) in the early 1980's samples as well as in the post-1995 samples from three different malaria-holoendemic regions were observed. In all post-1995 samples, one other predominant haplotype 7_8 as well as lower frequency haplotypes, unique to each location, were observed. These data indicate that during the early spread of CQR, a predominant msint2_msint3 haplotype swept across PNG under CQ selection, and now has independently accumulated diversity over time in different regions of PNG. These results are consistent with the results of a previous study [8], where significantly higher diversity was observed at the MS loci flanking pfcrt-SVMNT in the late 1990's samples than in the early 1980's samples. This suggests that although SVMNT remains the predominant CQR-associated pfcrt allele in PNG, its genetic background has accumulated significant diversity even under drug selection pressure.
Similar to the observations shown here, Vinayak et al. [31] found that a single major haplotype between pfcrt msint2_msint4 (AT_TA repeats; frequencies 0.222–0.879) was distributed across their six Indian sample collection sites. The frequencies of the minor haplotypes ranged from 0.02–0.244; 12 of the 32 minor haplotypes were distributed among sites separated by thousands of kilometers. In PNG, the major pfcrt msint2_msint3 haplotype (3_8) was present at all three sample collection sites at frequencies ranging from 0.667–0.955, while the 9 minor haplotypes were unique to each of the sites, and ranged in frequency from 0.011–0.048 (Table 4). Vinayak et al. [31] did not evaluate the polymorphism in msint3. Interestingly, when some of the same Indian samples were analysed for msint2_msint3 haplotypes, it was found that the same haplotype predominant in PNG (3_8) was also present in Indian samples at a frequency of 0.917 (n = 22/24). This observation suggests a relationship between PNG and Indian CQR P. falciparum strains that should be further evaluated. Finally, although Vinayak et al. [31] observed overall reduced msint2_msint4 haplotype diversity in lower malaria transmission areas, in PNG the lowest msint2_msint3 diversity was observed in the Liksul region (Madang area), which is known to experience holoendemic malaria transmission. As malaria ecology is known to be very different between PNG and India, it is possible that local factors influencing P. falciparum transmission dynamics contribute to some of the overall differences in frequency and distribution of intronic MS polymorphism between these two studies.
Recently, Ariey et al. used msint4 to look at the ancestry of CQR parasites at 16 survey sites throughout Africa [58]. By analysing the MS distribution in parasites carrying sensitive and resistant alleles, they reported a significant difference in the level of variability with 17 MS alleles present in the sensitive parasites vs. two MS alleles present in the resistant parasites [58]. Because 123 of the 125 resistant parasites from wide ranging survey sites carried the same MS allele, Ariey et al. concluded that Africa was invaded by a single CQR P. falciparum strain [58]. While it is clear from Ariey et al. [58] that a single intron-4 MS allele predominates among African CQR P. falciparum, results shown here and by Vinayak et al. [31] provide evidence that multiple intronic MS within pfcrt are polymorphic, and they have differing levels of variability. Therefore, to fully understand the ancestry, dispersal, and population genetics of CQR P. falciparum in Africa, or throughout the world, more complete surveillance of pfcrt intronic MS should be conducted. It would also be interesting to examine earlier sample sets to determine if parasites with a single pfcrt intronic MS haplotype invaded Africa, or if multiple haplotypes occurred and only one became predominant. Of further interest, given the recent reports that removal of CQ pressure in Africa is associated with a return of the wild-type pfcrt allele carrying strains and CQ sensitivity [59], analysis of polymorphisms in both intronic and flanking MS may provide useful insight regarding drug sensitivity distribution patterns.
Conclusion
In conclusion, highly polymorphic MS loci have been identified within the pfcrt gene. These new markers were used to reassess the relationships among P. falciparum in blood samples obtained from the early 1980's through 2003 from malaria-endemic sites in Madang and East Sepik Provinces of PNG. These results suggest that a single major haplotype associated with CQR has achieved widespread distribution in PNG, and that this sequence continues to generate new polymorphism within localized regions. Although the analysis shown here has focused largely on MS polymorphism associated with the common pfcrt-SVMNT allele, for the first time, the identification of the pfcrt-CVIET allele in PNG is reported. These CVIET parasites contain intronic MS haplotypes that are not present in the SVMNT parasites, suggesting that the CVIET parasites may have been imported from neighboring geographic regions. By using pfcrt intronic MS as CQ susceptibility markers, the continuing evolution of pfcrt can be monitored, the geographic and temporal patterns of CQS and CQR parasite populations can be further analysed, and relative fitness of CQS and CQR P. falciparum strains can be evaluated.
Authors' contributions
JTD and RKM performed the molecular typing of samples, data analysis, and interpretation, and wrote the manuscript. PM, IM, and JR collected and organised the PNG samples as well as developed the experimental design. YDS provided the Indian field samples and contributed to the experimental design. MS helped in data analysis and was involved in drafting the manuscript. PAZ was instrumental in the experimental design, data analysis, completion of the manuscript, and raising funds for this study.
Supplementary Material
Primers and PCR amplification conditions for pfcrt loci.
Ligase detection reaction primers for genotyping pfcrt codons.
Schematic of PCR primers used to characterise pfcrt intronic microsatellites.
Allele frequencies of intron-2 and intron-3 microsatellites in malaria-endemic regions of Papua New Guinea.
Acknowledgments
Acknowledgements
We thank BT Grimberg and DT McNamara for providing helpful comments on the manuscript. Also, we thank TA Smith and JC Wootton for critical commentary and advice during the course of this study. This work was supported by National Institutes of Health grant AI-52312.
Contributor Information
Jeana T DaRe, Email: jtd12@case.edu.
Rajeev K Mehlotra, Email: rkm@case.edu.
Pascal Michon, Email: pmichon@datec.net.pg.
Ivo Mueller, Email: pngimr_ivo@datec.net.pg.
John Reeder, Email: jreeder@burnet.edu.au.
Yagya D Sharma, Email: ydsharma_aiims@yahoo.com.
Mark Stoneking, Email: stoneking@eva.mpg.de.
Peter A Zimmerman, Email: paz@case.edu.
References
- Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- Trape JF. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg. 2001;64:12–17. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- Yung AP, Bennett NM. Chloroquine-resistant falciparum malaria in Papua New Guinea. Med J Aust. 1976;2:320–321. [PubMed] [Google Scholar]
- Grimmond TR, Donovan KO, Riley ID. Chloroquine resistant malaria in Papua New Guinea. P N G Med J. 1976;19:184–185. [PubMed] [Google Scholar]
- Cattani JA, Tulloch JL, Vrbova H, Jolley D, Gibson FD, Moir JS, Heywood PF, Alpers MP, Stevenson A, Clancy R. The epidemiology of malaria in a population surrounding Madang, Papua New Guinea. Am J Trop Med Hyg. 1986;35:3–15. doi: 10.4269/ajtmh.1986.35.3. [DOI] [PubMed] [Google Scholar]
- Dulay IS, Gibson FD, Eyeson-Annan MB, Narara A. Chloroquine resistance in Plasmodium falciparum and its geographical distribution in Papua New Guinea. P N G Med J. 1987;30:281–290. [PubMed] [Google Scholar]
- Mehlotra RK, Fujioka H, Roepe PD, Janneh O, Ursos LM, Jacobs-Lorena V, McNamara DT, Bockarie MJ, Kazura JW, Kyle DE, Fidock DA, Zimmerman PA. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci USA. 2001;98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Mattera G, Bhatia K, Reeder JC, Stoneking M, Zimmerman PA. Insight into the Early Spread of Chloroquine-Resistant Plasmodium falciparum Infections in Papua New Guinea. J Infect Dis. 2005;192:2174–2179. doi: 10.1086/497694. [DOI] [PubMed] [Google Scholar]
- Muller I, Bockarie M, Alpers M, Smith T. The epidemiology of malaria in Papua New Guinea. Trends Parasitol. 2003;19:253–259. doi: 10.1016/S1471-4922(03)00091-6. [DOI] [PubMed] [Google Scholar]
- al-Yaman F, Genton B, Mokela D, Narara A, Raiko A, Alpers MP. Resistance of Plasmodium falciparum malaria to amodiaquine, chloroquine and quinine in the Madang Province of Papua New Guinea, 1990–1993. P N G Med J. 1996;39:16–22. [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems TE, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci USA. 1991;88:3382–3386. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Kirkman LA, Fujioka H, Wellems TE. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/S0092-8674(00)80447-X. [DOI] [PubMed] [Google Scholar]
- Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV, Coulibaly D. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- Pillai DR, Labbe AC, Vanisaveth V, Hongvangthong B, Pomphida S, Inkathone S, Zhong K, Kain KC. Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. J Infect Dis. 2001;183:789–795. doi: 10.1086/318836. [DOI] [PubMed] [Google Scholar]
- Mayor AG, Gomez-Olive X, Aponte JJ, Casimiro S, Mabunda S, Dgedge M, Barreto A, Alonso PL. Prevalence of the K76T mutation in the putative Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene and its relation to chloroquine resistance in Mozambique. J Infect Dis. 2001;183:1413–1416. doi: 10.1086/319856. [DOI] [PubMed] [Google Scholar]
- Dorsey G, Kamya MR, Singh A, Rosenthal PJ. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J Infect Dis. 2001;183:1417–1420. doi: 10.1086/319865. [DOI] [PubMed] [Google Scholar]
- Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J Infect Dis. 2001;183:1535–1538. doi: 10.1086/320195. [DOI] [PubMed] [Google Scholar]
- Chen N, Russell B, Staley J, Kotecka B, Nasveld P, Cheng Q. Sequence polymorphisms in pfcrt are strongly associated with chloroquine resistance in Plasmodium falciparum. J Infect Dis. 2001;183:1543–1545. doi: 10.1086/320206. [DOI] [PubMed] [Google Scholar]
- Durand R, Jafari S, Vauzelle J, Delabre JF, Jesic Z, Le Bras J. Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol Biochem Parasitol. 2001;114:95–102. doi: 10.1016/S0166-6851(01)00247-X. [DOI] [PubMed] [Google Scholar]
- Goldberg DE, Slater AF, Cerami A, Henderson GB. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc Natl Acad Sci USA. 1990;87:2931–2935. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzekunov SM, Ursos LM, Roepe PD. Digestive vacuolar pH of intact intraerythrocytic P. falciparum either sensitive or resistant to chloroquine. Mol Biochem Parasitol. 2000;110:107–124. doi: 10.1016/S0166-6851(00)00261-9. [DOI] [PubMed] [Google Scholar]
- Saliba KJ, Folb PI, Smith PJ. Role for the Plasmodium falciparum digestive vacuole in chloroquine resistance. Biochem Pharmacol. 1998;56:313–320. doi: 10.1016/S0006-2952(98)00140-3. [DOI] [PubMed] [Google Scholar]
- Krogstad DJ, Gluzman IY, Kyle DE, Oduola AM, Martin SK, Milhous WK, Schlesinger PH. Efflux of chloroquine from Plasmodium falciparum: mechanism of chloroquine resistance. Science. 1987;238:1283–1285. doi: 10.1126/science.3317830. [DOI] [PubMed] [Google Scholar]
- Sanchez CP, McLean JE, Rohrbach P, Fidock DA, Stein WD, Lanzer M. Evidence for a pfcrt-associated chloroquine efflux system in the human malarial parasite Plasmodium falciparum. Biochemistry. 2005;44:9862–9870. doi: 10.1021/bi050061f. [DOI] [PubMed] [Google Scholar]
- Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, Cowman AF. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- Mu J, Ferdig MT, Feng X, Joy DA, Duan J, Furuya T, Subramanian G, Aravind L, Cooper RA, Wootton JC, Xiong M, Su XZ. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Nair S, Qin H, Singlam S, Brockman A, Paiphun L, Nosten F. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob Agents Chemother. 2005;49:2180–2188. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Johnson DJ, Fidock DA, Mungthin M, Lakshmanan V, Sidhu AB, Bray PG, Ward SA. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol Cell. 2004;15:867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Mittra P, Sharma YD. Wide variation in microsatellite sequences within each Pfcrt mutant haplotype. Mol Biochem Parasitol. 2006;147:101–108. doi: 10.1016/j.molbiopara.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Su X, Ferdig MT, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Su XZ, Roddam A, Day KP. Complex mutations in a high proportion of microsatellite loci from the protozoan parasite Plasmodium falciparum. Mol Ecol. 2000;9:1599–1608. doi: 10.1046/j.1365-294x.2000.01057.x. [DOI] [PubMed] [Google Scholar]
- Nair S, Williams JT, Brockman A, Paiphun L, Mayxay M, Newton PN, Guthmann JP, Smithuis FM, Hien TT, White NJ, Nosten F, Anderson TJ. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol Biol Evol. 2003;20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- Stoneking M, Jorde LB, Bhatia K, Wilson AC. Geographic Variation in Human Mitochondrial DNA from Papua New Guinea. Genetics. 1990;124:717–733. doi: 10.1093/genetics/124.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Lorry K, Kastens W, Miller SM, Alpers MP, Bockarie M, Kazura JW, Zimmerman PA. Random distribution of mixed species malaria infections in Papua New Guinea. Am J Trop Med Hyg. 2000;62:225–231. doi: 10.4269/ajtmh.2000.62.225. [DOI] [PubMed] [Google Scholar]
- Mehlotra RK, Kasehagen LJ, Baisor M, Lorry K, Kazura JW, Bockarie MJ, Zimmerman PA. Malaria infections are randomly distributed in diverse holoendemic areas of Papua New Guinea. Am J Trop Med Hyg. 2002;67:555–562. doi: 10.4269/ajtmh.2002.67.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Ziats MN, Bockarie MJ, Zimmerman PA. Prevalence of CYP2B6 alleles in malaria-endemic populations of West Africa and Papua New Guinea. Eur J Clin Pharmacol. 2006;62:267–275. doi: 10.1007/s00228-005-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara DT, Kasehagen LJ, Grimberg BT, Cole-Tobian J, Collins WE, Zimmerman PA. Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am J Trop Med Hyg. 2006;74:413–421. [PMC free article] [PubMed] [Google Scholar]
- Carnevale EP, Kouri D, DaRe JT, McNamara DT, Mueller I, Zimmerman PA. A Multiplex Ligase Detection Reaction-Fluorescent Microsphere Assay (LDR-FMA) for Simultaneous Diagnosis of Single Nucleotide Polymorphisms Associated with Plasmodium falciparum Drug Resistance. J Clin Microbiol. 2007;45:752–61. doi: 10.1128/JCM.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/S0031182099004552. [DOI] [PubMed] [Google Scholar]
- Excoffier LGL, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Anderson TJ. Mapping drug resistance genes in Plasmodium falciparum by genome-wide association. Curr Drug Targets Infect Disord. 2004;4:65–78. doi: 10.2174/1568005043480943. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, Chandramohan D, Sharp B. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361:1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Pearce R, Malisa A, Kachur SP, Barnes K, Sharp B, Roper C. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Mol Biol Evol. 2005;22:1834–1844. doi: 10.1093/molbev/msi177. [DOI] [PubMed] [Google Scholar]
- Nash D, Nair S, Mayxay M, Newton PN, Guthmann JP, Nosten F, Anderson TJ. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc Biol Sci. 2005;272:1153–1161. doi: 10.1098/rspb.2004.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Barry AE, Lyons EJ, Nielsen KM, Thomas SM, Choi M, Thakore SS, Day KP, Wirth DF, Hartl DL. Recent origin of Plasmodium falciparum from a single progenitor. Science. 2001;293:482–484. doi: 10.1126/science.1059878. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. The origin of malaria: mixed messages from genetic diversity. Nat Rev Microbiol. 2004;2:15–22. doi: 10.1038/nrmicro795. [DOI] [PubMed] [Google Scholar]
- Nielsen KM, Kasper J, Choi M, Bedford T, Kristiansen K, Wirth DF, Volkman SK, Lozovsky ER, Hartl DL. Gene conversion as a source of nucleotide diversity in Plasmodium falciparum. Mol Biol Evol. 2003;20:726–734. doi: 10.1093/molbev/msg076. [DOI] [PubMed] [Google Scholar]
- Kidgell C, Volkman SK, Daily J, Borevitz JO, Plouffe D, Zhou Y, Johnson JR, Le Roch K, Sarr O, Ndir O, Mboup S, Batalov S, Wirth DF, Winzeler EA. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2006;2:e57. doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagesha HS, Casey GJ, Rieckmann KH, Fryauff DJ, Laksana BS, Reeder JC, Maguire JD, Baird JK. New haplotypes of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene among chloroquine-resistant parasite isolates. Am J Trop Med Hyg. 2003;68:398–402. [PubMed] [Google Scholar]
- Huaman MC, Yoshinaga K, Suryanatha A, Suarsana N, Kanbara H. Short report: polymorphisms in the chloroquine resistance transporter gene in Plasmodium falciparum isolates from Lombok, Indonesia. Am J Trop Med Hyg. 2004;71:40–42. [PubMed] [Google Scholar]
- Ariey F, Fandeur T, Durand R, Randrianarivelojosia M, Jambou R, Legrand E, Ekala MT, Bouchier C, Cojean S, Duchemin JB, Robert V, Le Bras J, Mercereau-Puijalon O. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar J. 2006;5:34. doi: 10.1186/1475-2875-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers and PCR amplification conditions for pfcrt loci.
Ligase detection reaction primers for genotyping pfcrt codons.
Schematic of PCR primers used to characterise pfcrt intronic microsatellites.
Allele frequencies of intron-2 and intron-3 microsatellites in malaria-endemic regions of Papua New Guinea.