Abstract
NifS-like proteins provide the sulfur (S) for the formation of iron-sulfur (Fe-S) clusters, an ancient and essential type of cofactor found in all three domains of life. Plants are known to contain two distinct NifS-like proteins, localized in the mitochondria (MtNifS) and the chloroplast (CpNifS). In the chloroplast, five different Fe-S cluster types are required in various proteins. These plastid Fe-S proteins are involved in a variety of biochemical pathways including photosynthetic electron transport and nitrogen and sulfur assimilation. In vitro, the chloroplastic cysteine desulfurase CpNifS can release elemental sulfur from cysteine for Fe-S cluster biogenesis in ferredoxin. However, because of the lack of a suitable mutant allele, the role of CpNifS has not been studied thus far in planta. To study the role of CpNifS in Fe-S cluster biogenesis in vivo, the gene was silenced by using an inducible RNAi (interference) approach. Plants with reduced CpNifS expression exhibited chlorosis, a disorganized chloroplast structure, and stunted growth and eventually became necrotic and died before seed set. Photosynthetic electron transport and carbon dioxide assimilation were severely impaired in the silenced plant lines. The silencing of CpNifS decreased the abundance of all chloroplastic Fe-S proteins tested, representing all five Fe-S cluster types. Mitochondrial Fe-S proteins and respiration were not affected, suggesting that mitochondrial and chloroplastic Fe-S assembly operate independently. These findings indicate that CpNifS is necessary for the maturation of all plastidic Fe-S proteins and, thus, essential for plant growth.
Keywords: Fe-S proteins, inducible RNAi, photosynthesis, Arabidopsis thaliana
NifS-like proteins have cysteine desulfurase activity, which releases elemental sulfur (S) from the amino acid cysteine for the formation of iron-sulfur (Fe-S) clusters. First discovered in the nitrogen-fixing bacterium Azotobacter vinelandii (1), NifS-like proteins now represent a conserved group of proteins that are found in all of the domains of life (2). In bacteria, NifS-like proteins can be broadly classified on the basis of primary structure as either group I, which have a proposed general housekeeping role, or group II, which are likely required during oxidative stress. Escherichia coli contains 3 NifS-like proteins: IscS (group I), CsdA (group II), and SufS (group II) (for reviews see refs. 2 and 3). It is noteworthy that deletion of IscS is not lethal in E. coli; this mild phenotype is attributed to complementation by SufS (4).
In plants, Fe-S proteins are known to exist in the mitochondria, cytosol, and chloroplasts. The chloroplast contains five Fe-S cluster types: ferredoxin (Fd)-type 2Fe-2S, Rieske-type 2Fe-2S, 3Fe-4S, 4Fe-4S, and the siroheme 4Fe-4S (5, 6). These Fe-S clusters are used by an assortment of chloroplastic proteins that are involved in a diverse range of functions including protein import, sulfur and nitrogen reduction, chlorophyll synthesis, and photosynthetic electron transport (5).
Mitochondria and chloroplasts are thought to be the result of separate endosymbiotic events during the evolution of eukaryotes. Plants contain two distinct NifS-like proteins, one localized to mitochondria (MtNifS) (7) and the other localized to chloroplasts (CpNifS) (8, 9). MtNifS (group I) is most similar to bacterial IscS. The MtNifS homologue in yeast is required for Fe-S cluster formation in the mitochondria and cytosol, and MtNifS may function similarly in plants (7). CpNifS is a group II NifS-like protein (9). In vitro, CpNifS can provide the S for Fe-S cluster insertion into apo-Fd to form functional holo-Fd (10). Similar to the bacterial SufS, CpNifS cysteine desulfurase specific activity is low but greatly stimulated by a SufE protein (11, 12).
It may be hypothesized that CpNifS provides the sulfur for all five types of Fe-S proteins that occur in plastids. However, the specific function of CpNifS in chloroplastic Fe-S cluster assembly and its significance for plant development and survival has not been established in planta because of the lack of a suitable mutant. To determine the role of CpNifS in the synthesis of chloroplastic Fe-S clusters, we silenced the Arabidopsis CpNifS gene by using an ethanol-inducible RNAi construct and investigated the effects on levels of Fe-S proteins and photosynthesis.
Results
CpNifS Is Essential for Plant Growth and Maintenance of Chloroplast Structure.
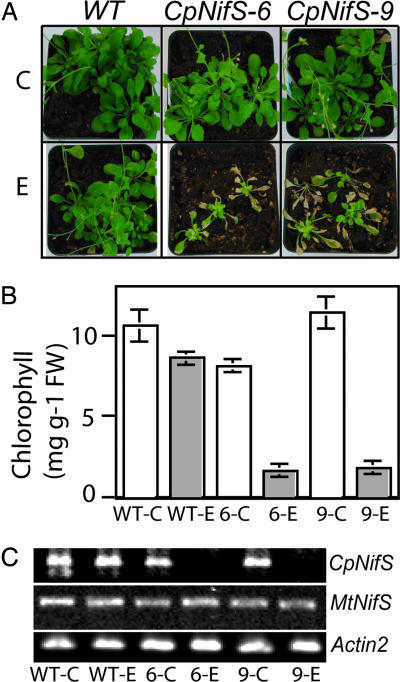
Initially, CpNifS was silenced constitutively by RNAi using the cauliflower mosaic virus 35S promoter. Lines in which CpNifS expression was significantly reduced displayed severely chlorotic cotyledons and died as seedlings. Although plants with milder phenotypes could be propagated, we found these constitutively driven CpNifS RNAi lines to be unstable for the trait. To avoid these problems, and to be able to study the effects of complete CpNifS silencing at a later developmental stage, an ethanol-inducible RNAi construct was used to silence CpNifS in Arabidopsis plants. Eleven transgenic inducible CpNifS RNAi lines were obtained and bred to homozygosity. Two of these lines, CpNifS-6 and CpNifS-9, were selected for further studies. To determine the efficacy of the inducible construct, wild-type (WT) and the two selected transgenic RNAi lines were grown for 2 weeks and then induced by ethanol treatment; control plants were not treated with ethanol. WT and transgenic plants that were not induced with ethanol typically did not show any signs of stress (Fig. 1A) and had normal development and seed production. Indeed, cytosolic ascorbate peroxidase I, a marker of oxidative stress in plants (13), was not induced in ethanol-treated plants [see supporting information (SI) Fig. 6]. Only in rare cases was slight chlorosis observed in uninduced transgenics, possibly because of leakiness of the ethanol-inducible promoter. After 3 weeks of ethanol treatment, the CpNifS-6 and CpNifS-9 transgenics showed severely stunted growth, chlorosis, and leaf necrosis. Ethanol-treated WT plants showed no such symptoms, albeit that ethanol-treated WT plants were sometimes slightly smaller after 3 weeks of treatment (Fig. 1A). At this stage, the leaves of ethanol-treated CpNifS-6 and CpNifS-9 plants had at least 5-fold lower chlorophyll content compared with WT and untreated control plants (Fig. 1B). When ethanol treatment was stopped at this stage, the CpNifS plants recovered and were able to set seed. However, continued ethanol treatment ultimately resulted in irreversible damage and death of the RNAi transgenics, whereas it had no visible or only a marginal effect on the WT.
Fig. 1.
Phenotypes of CpNifS-silenced plants. (A) Two-week old WT, CpNifS-6, and CpNifS-9 plants were treated with 2% ethanol (E) every 4 days for 3 weeks. Control plants (C) were treated with water. (B) Chlorophyll content in plants treated with ethanol or control plants. P < 0.05 for ethanol-treated CpNifS-6 and CpNifS-9. (C) CpNifS, MtNifS, and Actin2 transcript detection by RT-PCR 1 week after the start of ethanol treatment. FW, fresh weight.
To determine whether the stunted growth of ethanol-treated transgenic plants coincided with the loss of CpNifS mRNA, leaf samples were taken from treated and untreated plants at week 3 of plant growth (1 week after the start of ethanol treatment) for RT-PCR analysis. The CpNifS transcript was not detected in leaves of chlorotic CpNifS-6 and CpNifS-9 plants that were treated with ethanol (Fig. 1C). The mRNAs for Actin2 and MtNifS (NFS1) were not affected by ethanol treatment, showing that the RNAi ethanol-inducible construct is specific for the CpNifS gene product.
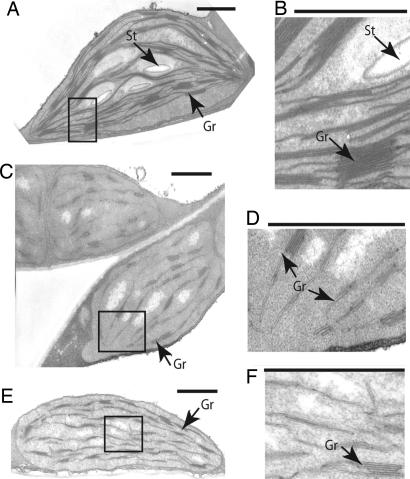
The severe chlorosis displayed by the ethanol-induced CpNifS-silenced plants prompted us to analyze their ultrastructure by transmission electron microscopy to determine the effects of CpNifS deficiency on leaf cell and plastid structure. The overall cell shape and the morphology of the nucleus appeared the same in ethanol-treated WT, CpNifS-6, and CpNifS-9 plants; it is also noteworthy that the structure of the mitochondria was unchanged in CpNifS plants, suggesting that MtNifS and mitochondrial processes were not affected (data not shown). However, chloroplast structure was drastically changed in plants in which CpNifS was silenced (Fig. 2). Compared with the discrete and stacked thylakoid membrane grana displayed by WT, the grana in silenced CpNifS-6 and CpNifS-9 plants were hypertrophied and dissociated from each other. Also of interest was the absence of starch granules in CpNifS-6 and CpNifS-9, likely caused by a disruption of photosynthesis.
Fig. 2.
Chloroplast ultrastructure is altered in CpNifS-silenced plants. Leaf samples were fixed 10 days after ethanol treatment of WT (A and B), CpNifS-6 (C and D), and CpNifS-9 (E and F) plants. Thin sections were examined by transmission electron microscopy. B, D, and F show magnifications of the boxed areas in A, C, and E, respectively. St, starch granules; Gr, grana. (Scale bars, 1 μm.)
Taken together, these results strongly suggest that CpNifS is an essential protein in Arabidopsis. CpNifS loss of function causes pleiotropic phenotypes and eventually plant death. To analyze the primary cause of these phenotypes and to insight into the direct function of CpNifS in plants before pleiotropic phenotypes were apparent, further experiments with the inducible RNAi lines were performed by using plants in which ethanol treatment started 3 weeks after germination, analyzing the plants 10 days after induction.
CpNifS Mutants Have Impaired Photosynthesis but Unaltered Respiration.
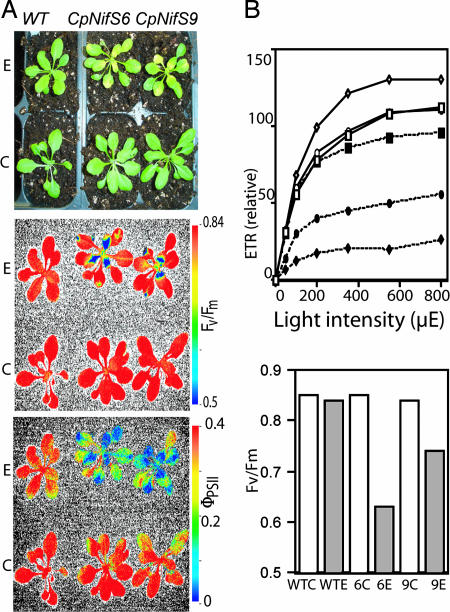
We hypothesized that CpNifS silencing would affect photosynthesis, because photosynthetic electron transport requires many Fe-S proteins, particularly in photosystem I (PSI). To determine how loss of CpNifS affects photosynthetic electron transport in intact plants, a chlorophyll fluorescence imaging system was used. For silenced plants and controls, images were captured of two chlorophyll fluorescence parameters: Fv/Fm, which indicates the maximum photochemical efficiency (intactness) of PSII, and ΦPSII, which indicates the fraction of PSII complexes available for photochemistry and indirectly measures the flux of electrons out of PSII (14).
Transgenic plants that were ethanol-treated showed chlorosis, particularly in younger leaves (Fig. 3A). However, the older leaves that had already fully expanded before induction had not become chlorotic at this stage (10 days after induction). Accordingly, when treated with ethanol, Fv/Fm was strongly reduced in the younger leaves of CpNifS-6 and CpNifS-9 transgenics compared with WT and untreated controls, but older leaves were less affected (Fig. 3A). The electron flow, ΦPSII, was reduced in both the young and the older leaves of the transgenics, compared with WT.
Fig. 3.
Chlorophyll fluorescence analysis. (A) Chlorophyll fluorescence imaging. Shown are a regular-color photograph (Top) and false-color images for Fv/Fm (Middle) and ΦPSII (Bottom) of the same plants. False-color scales for fluorescence parameters are shown to the right (red color represents the highest values and blue represents the lowest values). (B Upper) Relative ETR at varying light intensities. Squares, diamonds, and circles represent WT, CpNifS-6, and CpNifS-9, respectively. Open symbols correspond to untreated controls, and closed symbols signify ethanol treatment. (B Lower) Fv/Fm. Note: standard errors were too small to be plotted. P < 0.05 for ethanol-treated CpNifS-6 and CpNifS-9. C, control plants; E, ethanol-treated plants.
To gain more quantitative insight into how CpNifS silencing affects photosynthesis, ΦPSII was measured and used to estimate the electron-transport rate (ETR) over a range of light intensities in expanded leaves in which chlorosis was absent or minimal. At all light intensities the ΦPSII and, as a consequence, ETR were reduced; ETR saturated at much lower light intensities in CpNifS-6 and CpNifS-9 plants induced with ethanol, compared with WT and untreated plants (Fig. 3B). The reduced ETR correlated with a deficiency in CpNifS mRNA in the same leaves (Fig. 1C). The CpNifS-silenced plants also showed a decrease in Fv/Fm (Fig. 3C), suggesting photoinhibition or some other effect that reduced the maximum photochemical efficiency of PSII.
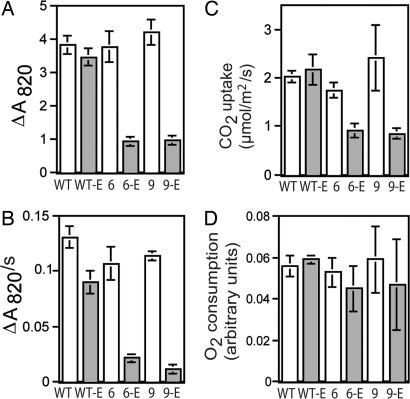
All of the Fe-S proteins involved in photosynthetic electron transport are found downstream of PSII: one 2Fe-2S cluster is in the Rieske protein of the cytochrome b6f complex, and three 4Fe-4S clusters are found in PSI (5). The photochemical activity of PSI was analyzed in leaf disks by measuring the light-induced absorbance change at 820 nm (ΔA820), which occurs with photooxidation of the P700 reaction center of PSI (15, 16). Ethanol-treated CpNifS-6 and CpNifS-9 plants had less than one third of the ΔA820 exhibited by ethanol-treated WT or untreated plants (Fig. 4A), which indicates a substantial loss of photochemically active P700. The rate of dark reduction of P700 after the oxidizing flash was also reduced in the CpNifS-silenced plants, indicating that the flow of electrons into PSI from upstream donors was slower (Fig. 4B). To test whether the reduced electron-transport activity limited plant productivity, we measured photosynthetic CO2 fixation. Indeed, the CO2 fixation rate in the light was reduced by 50% in the ethanol-treated CpNifS-6 and CpNifS-9 plants compared with WT and untreated plants (Fig. 4C). However, oxygen consumption in the dark, which is indicative of mitochondrial respiration, was not affected (Fig. 4D). Thus, CpNifS silencing does not appear to disrupt mitochondrial function, while severely affecting chloroplast function.
Fig. 4.
PSI activity and CO2 assimilation. WT, 6, and 9 indicate control plants for WT, CpNifS-6, and CpNifS-9 respectively, and WT-E, 6-E, and 9-E indicate the respective ethanol-treated plants. (A) PSI activity. Shown is the extent of the ΔA820 (absorbance change at 820 nm, indicative of the P700 activity of PSI) induced with a flash of saturating light, in relative units. (B) Rate of reduction of PSI by upstream electron donors. Values, in relative units, represent the initial rate of the absorbance change at 820 nm (ΔA820/s) in darkness after complete photooxidation of P700, which results mainly from reduction of P700+ by upstream electron donors. (C) Leaf photosynthetic CO2 uptake. (D) Leaf respiratory O2 consumption. P < 0.05 for ethanol-treated 6-E and 9-E for A–C. All values are the average and standard error of five independent measurements.
Levels of Chloroplastic Fe-S Proteins Are Reduced by CpNifS Silencing.
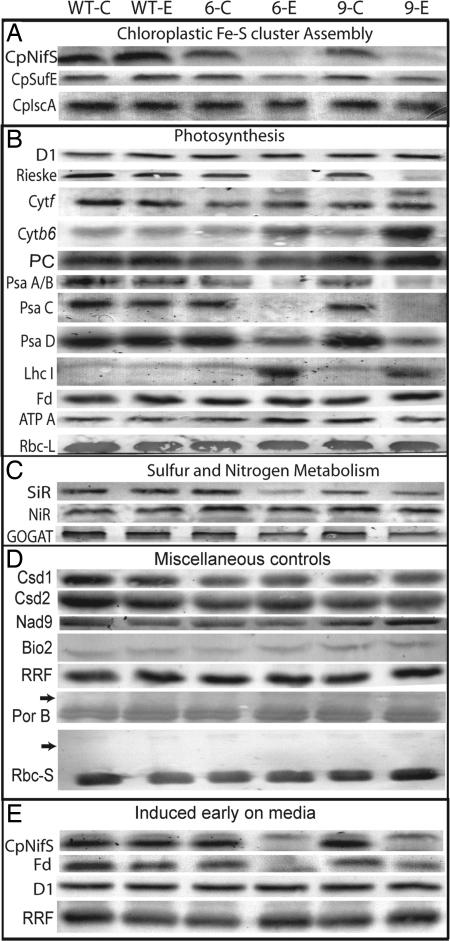
We next investigated the direct effect of CpNifS silencing on the abundance of chloroplast proteins with or without Fe-S clusters (Fig. 5A). Consistent with an absence of CpNifS mRNA, very little CpNifS protein was detected in transgenic plants treated with ethanol. The abundance of the CpNifS activator, CpSufE, was decreased in the absence of CpNifS, compared with WT and untreated controls. In contrast, the relative abundance of CpIscA, a putative scaffold protein for chloroplastic Fe-S cluster formation (17), was hardly affected by CpNifS silencing.
Fig. 5.
Effect of CpNifS silencing on protein levels. Immunoblot analysis for selected proteins is shown. Lanes labeled WT-C, 6-C, and 9-C indicate control plants for WT, CpNifS-6, and CpNifS-9 respectively, and lanes labeled WT-E, 6-E, and 9-E indicate the respective ethanol-treated plants. (A–D) Samples from plants grown on soil either under control conditions or after treatment with 2% ethanol for 10 days: (A) Chloroplastic Fe-S cluster assembly; (B) photosynthesis; (C) sulfur and nitrogen metabolism; (D) miscellaneous controls. (E) Protein extracts from plants grown from germination on 0.5 Murashige and Skoog medium + 1% sucrose plates with or without ethanol. Total protein extracts (25 μg of protein) were separated by SDS/PAGE and blotted to nitrocellulose before immunostaining with the indicated antibodies. RRF was used as a loading control. D1, D1 subunit of PSII; PC, plastocyanin; ATP A, ATP synthase subunit A; LhcI, light-harvesting complex proteins of PSI; Rbc-L, large subunit of Rubisco; Csd, Cu/Zn superoxide dismutase; Nad9, subunit 9 of mitochondrial complex 1; Bio2, mitochondrial biotin synthase; PsaA–D, PSI subunits A–D; Por B, protochlorophyllide oxidoreductase; Rbc-S, small subunit of Rubisco. Specific bands were identified by immunostaining and by their molecular weight. Arrows indicate where precursor proteins would have been predicted to accumulate. For each protein, the relevant section of one representative blot of three independent repeats is shown.
To identify which specific steps of photosynthesis may be affected by the silencing of CpNifS, we examined the abundance of a variety of proteins involved in photosynthesis (Fig. 5B). CpNifS silencing did not affect the D1 protein, a component of the PSII reaction center in which no Fe-S clusters are present (18), suggesting that most PSII was intact. In contrast, components of the cytochrome b6f complex were affected by CpNifS silencing. Most noteworthy is the near absence of the 2Fe-2S Rieske protein. Although the abundance of cytochrome f was not affected, cytochrome b6 was more abundant in CpNifS-silenced plants. Both cytochrome f and cytochrome b6 are heme (Fe) proteins and chloroplast-encoded. A very profound effect of CpNifS silencing was observed in PSI. PsaA, PsaB, and PsaC, all 4Fe-4S proteins, were strongly reduced or absent in CpNifS-silenced plants. The PsaD protein, which is not an Fe-S cluster protein but thought to be involved in Fd docking (19), was reduced also. This reduction in PsaD abundance may reflect the overall lack of integrity of PSI in CpNifS-silenced plants. Perhaps to compensate for the very low abundance of PSI reaction-center proteins in CpNifS-silenced plants, there was an increased abundance of subunits of the light-harvesting complex proteins of PSI (LhcI).
Surprisingly, no effect was seen on the abundance of the 2Fe-2S protein Fd in soil-grown CpNifS-silenced plants. One possible explanation is that the Fd already present in the 3-week-old plants before ethanol induction was very stable, so that Fd levels did not go down once further production of holo-Fd ceased. To test this possibility, we induced CpNifS silencing during germination on media plates. After 1 week, ethanol-treated transgenic plants had extreme chlorosis and stunted growth compared with WT and untreated plants. Western blotting showed that CpNifS and Fd were absent in these ethanol-treated transgenics, indicating that Fd levels do decrease if CpNifS is silenced at an early stage in plant development (Fig. 5E). Therefore, holo-Fd may be very stable and remain abundant even when the production of its cofactor is impeded. D1, a plastid- encoded subunit of PSII, and ribosome-recycling factor (RRF), which is a nuclear-encoded plastid protein, were not affected by CpNifS silencing from germination.
In the soil-grown plants that were ethanol-induced at the 3-week-old stage, the ATP-A subunit of chloroplastic ATP synthase and the large subunit of ribulose bisphosphate carboxylase oxygenase (Rubisco), involved in photosynthetic ATP production and CO2 fixation, respectively, were not affected; neither of these are Fe-S proteins (Fig. 5B). There was also no major effect on plastocyanin, the copper protein that is active in electron transport between cytochrome b6f and PSI (Fig. 5B). Two other copper-containing proteins, Cu/ZnSOD1 and Cu/ZnSOD2, involved in free radical scavenging, were also not affected by CpNifS silencing (Fig. 5D).
In addition to the photosynthetic proteins described above, other plastidic Fe-S cluster proteins are expected to depend on CpNifS for their maturation. Indeed, the 3Fe-4S cluster protein Fd-glutamine-2-oxoglutarate aminotransferase (GOGAT) was slightly less abundant compared with WT and untreated controls. Moreover, sulfite reductase (SiR) and, to a lesser extent, nitrite reductase (NiR) were reduced in CpNifS-silenced plants (Fig. 5C). SiR and NiR contain the siroheme 4Fe-4S cluster type unique to plants. The polypeptide level of NiR was reduced only moderately (30%) compared with WT and untreated plants. In contrast, the enzyme activity of NiR was significantly reduced, by 70% in CpNifS-6 and 45% in CpNifS-9, compared with untreated transgenic plants (SI Fig. 7). The observation that the activity of NiR was reduced to a greater extent than NiR protein levels suggests that apo-NiR, lacking the Fe-S cofactor, had accumulated in CpNifS-deficient plants.
The levels of CpNifS and several Fe-S proteins were monitored over a 10-day period after ethanol induction to determine how quickly CpNifS decreased in CpNifS-6 plants and how quickly it was followed by a decrease in chloroplastic Fe-S proteins. CpNifS began to diminish in transgenic plants 1 day after ethanol induction and had almost completely disappeared by day 10 (SI Fig. 8). Protein abundance of PSI subunits PsaA/B began to decrease on day 2 after ethanol induction, whereas decline of PsaC levels started on day 7. By day 10, a slight reduction in GOGAT, NiR, and SiR was seen. Levels of the control protein RRF remained constant over the course of 10 days.
To determine whether the decrease in chloroplastic Fe-S proteins was caused by a decrease in mRNA, a Northern blot was performed to detect the transcript abundance of Rieske, GOGAT, and SiR. A decrease in mRNA was not observed (SI Fig. 6) despite a large reduction in Rieske, GOGAT, and SiR protein levels (Fig. 5 B and C), which suggests that these Fe-S proteins need their appropriate cofactor to be stable and supports the hypothesis that CpNifS is critical for Fe-S cluster formation in the chloroplast.
In view of the decrease in several abundant Fe-S proteins in the CpNifS-silenced lines, leaf nutrient status was investigated (SI Table 1). We did not see a reduction in total leaf iron content on the basis of dry mass; in fact, a modest increase (+25%, nonsignificant) was seen in the ethanol-induced transgenics, compared with ethanol-induced WT. Sulfur and phosphorous levels were significantly increased (by 25% and 50%, respectively) in these same plants.
To rule out the possibility that CpNifS plants suffered from a defective chloroplast-import machinery, protein levels of the nuclear encoded protochlorophyllide reductase B and the small subunit of Rubisco were tested to determine whether their mature protein levels are reduced together with accumulation of precursors, as reported for chloroplast-import mutants (20, 21). Both proteins were present in mature size at equal levels in all plant types and treatments. Protochlorophyllide reductase B and the small subunit of Rubisco precursor-sized proteins did not accumulate, suggesting that protein import was not affected by CpNifS silencing. Finally, the abundance of two mitochondrial Fe-S proteins, Nad9 (a component of respiratory complex I) and biotin synthase, were not affected by CpNifS silencing (Fig. 5D). In summary, CpNifS silencing seems to specifically affect the maturation of Fe-S proteins in plastids.
Discussion
Silencing of CpNifS severely affected levels of chloroplastic Fe-S proteins and photosynthesis, and prolonged silencing resulted in a pleiotropic-stressed phenotype and eventually plant death. These results suggest that CpNifS is an essential protein that functions in plastid Fe-S cluster assembly and cannot be bypassed or complemented by MtNifS. When silencing was induced after the seedling stage, specific and reversible defects could be observed that yielded information about the functions of CpNifS. Silencing of CpNifS caused a defect in the accumulation of all eight chloroplastic Fe-S cluster proteins that were tested. The Fe-S proteins affected by CpNifS silencing together represent all five types of Fe-S clusters found in plastids, supporting the hypothesis that the cysteine desulfurase activity of CpNifS is required for the maturation of all Fe-S proteins in this organelle. Two mitochondrial Fe-S proteins were not affected, lending evidence that Fe-S cluster assembly in the mitochondria can operate independently of the chloroplastic cysteine desulfurase.
Fd did not exhibit any decrease when ethanol was initiated at week 3. However, Fd was absent when CpNifS silencing was induced with ethanol from germination, suggesting that holo-Fd is very stable once formed. In contrast to Fd, several other chloroplastic Fe-S proteins had decreased significantly 10 days after CpNifS silencing at week 3. These different delays in reduction of Fe-S proteins after CpNifS silencing may reflect protein stability or priority of Fe-S cofactor delivery.
The observed defects in photosynthetic electron transport and carbon fixation likely were a consequence of the lack of thylakoid Fe-S proteins, particularly in the cytochrome b6f complex and PSI. Indeed, PSI function was severely compromised after CpNifS silencing. At the same time, PSII was only marginally affected in comparison, as evidenced by the presence of the D1 protein and functional heat dissipation, measured as nonphotochemical quenching of chlorophyll fluorescence (data not shown). A comparison of the ΦPSII and Fv/Fm images suggests that ΦPSII was affected before Fv/Fm, which may imply that damage to PSII could be a secondary consequence of a downstream defect in photosynthetic electron transport.
The altered chloroplast ultrastructure observed in CpNifS-silenced lines is reminiscent of the ultrastructure reported for an APO1 mutant (22) that affects PSI accumulation, as well as a mutant in Hcf101 and other mutations that affect PSI (23, 24). Therefore, the dilated stromal lamellae and absence of grana may be a consequence of a lack of PSI.
Mitochondria and chloroplasts originated from separate endosymbiotic events during the evolution of eukaryotes, and the two organelles have separate NifS-like proteins. It is likely that these two NifS-like proteins with their different properties each evolved to function optimally in their respective environments. The main function of mitochondria is to carry out the oxygen-consuming process of respiration, whereas chloroplasts perform the oxygen-generating process of photosynthesis. Thus, although both organelles contain an electron-transport chain that depends on Fe-S protein assembly, they contrast in redox conditions. Moreover, photosynthesis is known to produce reactive oxygen species, which can lead to oxidative stress. Fe-S cluster biosynthesis is particularly sensitive to oxygen. Therefore, it is not surprising that the chloroplastic NifS that has to operate under high-oxygen conditions is most similar to the bacterial SufS, which is thought to function under oxidative stress (3). MtNifS is most similar to bacterial IscS, the housekeeping NifS-like protein that is more sensitive to oxygen. Therefore, MtNifS likely would not function properly in an oxygen-producing compartment.
Another difference between the chloroplast and mitochondrion is that the chloroplast is the main site of cysteine synthesis in plant cells. It may be important to tightly control the cysteine desulfurase activity of the chloroplastic NifS, to avoid futile cycling. CpNifS may be particularly suited for the chloroplast because its cysteine desulfurase activity is extremely low in the absence of its activator CpSufE, in contrast to group I NifS-like proteins such as IscS and MtNifS (11). In summary, the two Fe-S cluster biosynthesizing machineries in the chloroplast and mitochondrion likely have different evolutionary origins and display properties that fit their function and environmental conditions.
After these two Fe-S biosynthesis machineries came together in the same plant cell, have they shared or transferred some of their functions? As shown here, CpNifS silencing is lethal in Arabidopsis and affects all five chloroplast Fe-S cluster types. Thus, MtNifS cannot complement the function of CpNifS in the biogenesis of any of these cluster types. At this point, it has not been reported whether MtNifS is essential as well, as was shown to be the case in yeast (25). In our studies, CpNifS silencing had no effects on mitochondrial Fe-S protein levels or respiration, suggesting that the mitochondrial Fe-S biogenesis machinery does not depend on CpNifS. Together, these results indicate that in plants, mitochondria and chloroplasts still have separate, essential cysteine desulfurases and Fe-S cluster assembly machineries.
Materials and Methods
Generation of CpNifS-Silencing Constructs and Induction.
Standard cloning techniques were used to make the plant-transformation constructs and to generate transgenic Arabidopsis thaliana. For a detailed description of the cloning steps, see SI Text, Supporting Information on the Cloning and Plant Transformation. The primers used in plasmid construction and verification are listed in SI Table 2. Ten constitutive RNAi lines and 11 inducible RNAi lines were obtained, which were selfed and propagated to homozygosity. Two of these lines were used for further functional characterization: CpNifS-RNAi-6 and CpNifS-RNAi-9 (denoted as CpNifS-6 and CpNifS-9, respectively). To induce the RNAi construct, plants grown in soil were sprayed and soil-drenched every 4 days with a 2% ethanol solution, a concentration that was reported not to induce stress (26, 27); untreated control plants were sprayed with water. For RNAi induction on agar medium, plants were germinated on 0.5 strength Murashige and Skoog medium + 1% sucrose (28) solidified with 0.4% Agargel (Sigma) in 15-cm-diameter Petri dishes with 50 μl of 100% ethanol (or water for controls) placed in the center at the time of germination.
RT-PCR and Immunoblotting.
The presence of transcripts in plants was detected by using RT-PCR (29). Protein extraction, SDS/PAGE, and immunoblot analysis were performed essentially as described (30). Leaf tissue for protein analysis was collected 10 days after induction. Antibodies for CpNifS (9), CpSufE (11), CpIscA (17), Fd and SiR (31), cytochrome f, light-harvesting complex of PSI, and PsaA/B (32), Fd-GOGAT and NiR (33), cytochrome b6 and the Rieske subunit (34), PsaC and PsaD (35), protochlorophyllide reductase B and the small subunit of Rubisco (36), and the chloroplastic RRF (37) have been described. Specific antibodies for the D1 subunit of PSII and subunit A of ATP synthase were generous gifts from Alice Barkan (University of Oregon, Eugene) and Anna Sokolenko (Ludwig-Maximilians University, München, Germany), respectively. Specific antibodies for mitochondrial biotin synthase (38) and Nad9 (39) have been described. The intensity of mRNA and protein bands was quantified by using Image J imaging software [National Institutes of Health, Bethesda (http://rsb.info.nih.gov/ij)].
Electron Microscopy.
Leaves were sampled from soil-grown plants 10 days after the start of ethanol treatment. Fixation and sectioning before analysis by transmission electron microscopy were performed as described (40).
Photosynthesis and Respiration Measurements.
Chlorophyll content was assayed as described (41). Chlorophyll fluorescence images of Fv/Fm and ΦPSII were captured from control and ethanol-treated dark-adapted soil-grown plants by using a Photon System Instruments imaging system (Photon System Instruments, Brno, Czech Republic). Default protocol settings were used with an actinic light intensity of 110 μE. A fluorescence monitoring system chlorophyll fluorometer (Hansatech, Cambridge, U.K.) was used for quantitative chlorophyll fluorescence analysis on detached, fully expanded leaves taken from dark-adapted plants. Fv/Fm, ΦPSII, and ETR were calculated as described (14).
Photooxidation and dark-reduction kinetics of P700 (PSI) were measured in leaf disks by determining the light-induced absorbance change at 820 nm (ΔA820) (16, 42) and at a saturating light intensity of 1600 μE, determined empirically. Carbon assimilation was assayed in detached leaves at 770 μE by using a Qubit Systems analyzer according to manufacturer instructions (Qubit Systems, Kingston, Ontario, Canada). Oxygen consumption by dark respiration was monitored in leaf tissue with a Hansatech LD2/3 leaf-disk O2 electrode system maintained at 26°C.
Elemental Analysis, Enzyme Activity, and Statistics.
Elemental composition was measured as described previously (43). Enzyme activity of NiR was measured according to ref. 44. All statistical analyses (ANOVA, t tests) were performed by using the Jmp-In software package (SAS Institute, Cary, NC).
Supplementary Material
Acknowledgments
We acknowledge Syngenta for supplying the ethanol-inducible construct vectors. We are also grateful to the following colleagues for kindly providing antibodies used in this study: Drs. Iwona Adamska, Alice Barkan, Geraldine Bonnard, Antonio Marquez Cabeza, Toshiharu Hase, Norbert Rolland, Henrik Sheller, and Anna Sokolenko. This work was supported by U.S. Department of Agriculture grants USDA-NRI 2003-35318-13758 and 2005-35318-16212 (to M.P. and E.A.H.P.-S).
Abbreviations
- Fd
ferredoxin
- MtNifS
mitochondrial NifS-like protein
- CpNifS
chloroplastic NifS-like protein
- PSI/II
photosystem I/II
- ETR
electron-transport rate
- RRF
ribosome-recycling factor
- Rubisco
ribulose bisphosphate carboxylase oxygenase
- SiR
sulfite reductase
- NiR
nitrite reductase
- GOGAT
2-oxoglutarate aminotransferase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700774104/DC1.
References
- 1.Zheng L, Cash CL, Flint DH, Dean DR. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DC, Dean DR, Smith AD, Johnson MK. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 3.Mihara H, Esaki N. Appl Microbiol Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi Y, Tokumoto U. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 5.Balk J, Lobréaux S. Trends Plant Sci. 2005;10:324–331. doi: 10.1016/j.tplants.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Ye H, Pilon M, Pilon-Smits EAH. New Phytol. 2006;171:285–292. doi: 10.1111/j.1469-8137.2006.01751.x. [DOI] [PubMed] [Google Scholar]
- 7.Kushnir S, Babiychuk E, Storozhenko S, Davey MW, Papenbrock J, De Rycke R, Engler G, Stephan UW, Lange H, Kispal G, et al. Plant Cell. 2001;13:89–100. doi: 10.1105/tpc.13.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leon S, Touraine B, Briat JF, Lobréaux S. Biochem J. 2002;366:557–564. doi: 10.1042/BJ20020322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pilon-Smits EAH, Garifullina GF, Abdel-Ghany S, Kato S, Mihara H, Hale KL, Burkhead JL, Esaki N, Kurihara T, Pilon M. Plant Physiol. 2002;130:1309–1318. doi: 10.1104/pp.102.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye H, Garifullina GF, Abdel-Ghany SE, Zhang L, Pilon-Smits EAH, Pilon M. Planta. 2005;220:602–608. doi: 10.1007/s00425-004-1388-1. [DOI] [PubMed] [Google Scholar]
- 11.Ye H, Abdel-Ghany SE, Anderson TD, Pilon-Smits EAH, Pilon M. J Biol Chem. 2006;281:8958–8969. doi: 10.1074/jbc.M512737200. [DOI] [PubMed] [Google Scholar]
- 12.Xu XM, Møller SG. EMBO J. 2006;25:900–909. doi: 10.1038/sj.emboj.7600968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxwell K, Johnson GN. J Exp Bot. 2000;359:659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- 15.Klughammer C, Schreiber U. Z Naturforsch [C] 1991;46:233–244. [Google Scholar]
- 16.Martin RE, Thomas DJ, Tucker DE, Herbert SK. Plant Cell Environ. 1997;20:1451–1461. [Google Scholar]
- 17.Abdel-Ghany SE, Ye H, Garifullina GF, Zhang L, Pilon-Smits EAH, Pilon M. Plant Physiol. 2005;138:161–172. doi: 10.1104/pp.104.058602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raven JA, Evans MC, Korb RE. Photosynth Res. 1999;60:111–149. [Google Scholar]
- 19.Lagoutte B, Hanley J, Bottin H. Plant Physiol. 2001;126:307–316. doi: 10.1104/pp.126.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, Kessler F, Schnell DJ. Plant Cell. 2005;17:1482–1496. doi: 10.1105/tpc.105.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- 22.Amann K, Lezhneva L, Wanner G, Herrmann RG, Meurer J. Plant Cell. 2004;16:3084–3097. doi: 10.1105/tpc.104.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lezhneva L, Amann K, Meurer J. Plant J. 2004;37:174–185. doi: 10.1046/j.1365-313x.2003.01952.x. [DOI] [PubMed] [Google Scholar]
- 24.Stockel J, Bennewitz S, Hein P, Oelmuller R. Plant Physiol. 2006;141:870–878. doi: 10.1104/pp.106.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lill R, Kispal G. Trends Biochem Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- 26.Salter MM, Paine JA, Riddell KV, Sepson I, Greenland AJ, Caddick MX, Tomsett AB. Plant J. 1998;16:127–132. doi: 10.1038/nbt0298-177. [DOI] [PubMed] [Google Scholar]
- 27.Roslan HA, Salter MG, Wood CD, White MRH, Craft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al. Plant J. 2001;28:225–235. doi: 10.1046/j.1365-313x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- 28.Murashige T, Skoog F. Physiol Plant. 1962;15:437–497. [Google Scholar]
- 29.Schiavon M, Zhang L, Abdel-Ghany SE, Pilon M, Malagoli M, Pilon-Smits EAH. Physiol Plant. 2007;129:342–350. [Google Scholar]
- 30.Rensink WA, Pilon M, Weisbeek P. Plant Physiol. 1998;118:691–699. doi: 10.1104/pp.118.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonekura-Sakakibara K, Onsa Y, Ashikari T, Tanaka Y, Kusumi T, Hase T. Plant Physiol. 2000;122:887–894. doi: 10.1104/pp.122.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson U, Heddad M, Adamska I. Plant Physiol. 2003;132:811–820. doi: 10.1104/pp.102.019281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pajuelo E, Borrero JA, Márquez AJ. Plant Sci. 1993;95:9–21. [Google Scholar]
- 34.Alt J, Westhoff P, Sears BB, Nelson N, Hurt E, Hauska G, Herrmann RG. EMBO J. 1983;2:979–986. doi: 10.1002/j.1460-2075.1983.tb01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knoetzel J, Mant A, Haldrup A, Jensen PE, Sheller HV. FEBS Lett. 2002;510:145–148. doi: 10.1016/s0014-5793(01)03253-7. [DOI] [PubMed] [Google Scholar]
- 36.Philippar K, Geis T, Ilkavets I, Oster U, Schwenkert S, Meurer J, Soll J. Proc Natl Acad Sci USA. 2007;104:678–683. doi: 10.1073/pnas.0610062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolland N, Janosi L, Block MA, Shuda M, Teyssier E, Miège C, Chéniclet C, Carde JP, Kaji A, Joyard J. Proc Natl Acad Sci USA. 1999;96:5464–5469. doi: 10.1073/pnas.96.10.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldet B, Alban C, Douce R. FEBS Lett. 1997;419:206–210. doi: 10.1016/s0014-5793(97)01458-0. [DOI] [PubMed] [Google Scholar]
- 39.Lamattina L, Gonzalez D, Gualberto JM, Grienenberger JM. Eur J Biochem. 1993;217:831–838. doi: 10.1111/j.1432-1033.1993.tb18311.x. [DOI] [PubMed] [Google Scholar]
- 40.Antunes SM, Ha SB, Tewari-Singh N, Morey KJ, Trofka AM, Kugrens P, Deyholos M, Medford JI. Plant Biotechnol J. 2006;4:605–622. doi: 10.1111/j.1467-7652.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 41.Porra RJ, Thompson WA, Kriedemann PE. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- 42.Herbert SK, Martin RE, Fork DC. Photosynth Res. 1995;46:277–285. doi: 10.1007/BF00020441. [DOI] [PubMed] [Google Scholar]
- 43.Pilon-Smits EAH, Hwang S, Lytle CM, Zhu Y, Tai JC, Bravo RC, Chen Y, Leustek T, Terry M. Plant Physiol. 1999;119:123–132. doi: 10.1104/pp.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi M, Sasaki Y, Morikawa H. Plant Physiol. 2001;126:731–741. doi: 10.1104/pp.126.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.