Abstract
Green tea polyphenol, epigallocatechin-3-gallate (EGCG) differentially regulates the cellular growth of cancer cells in a p53-dependent manner through apoptosis and/or cell cycle arrest. In an effort to further elucidate the mechanism of differential growth regulation by EGCG, we have investigated the role of the tyrosine phosphatase, SHP-2. Comparing the responses of mouse embryonic fibroblasts (MEFs), expressing either WT or functionally inactive/truncated SHP-2, we find that inactivation of SHP-2 remarkably sensitizes cells to EGCG-mediated killing. MEFs lacking functional SHP-2 undergo massive apoptosis upon treatment with EGCG. By comparing gene expression profiles, we have identified a set of transcriptional targets of p53 that are differentially modulated in cells undergoing apoptosis. Western blot and real-time PCR analyses of a select group of genes further confirm that the expression is SHP-2-dependent. Similar observations were made in MEFs lacking p53, confirming that the expression of these “p53 target genes” is p53-independent. In addition, EGCG treatment induced the expression of p73 mRNA and protein in both cell types, but not p63. Inactivation of p73 in cells expressing nonfunctional SHP-2 markedly inhibited apoptosis and p53 target gene expression. Although phosphorylation of JNK is differentially regulated by SHP2, it was found to be dispensable for EGCG-induced apoptosis and p53 target gene expression. Our results have identified SHP-2 as a negative regulator of EGCG-induced-apoptosis and have identified a subset of p53 target genes whose expression is paradoxically not mediated by p53 but by one of its family members, p73.
Keywords: green tea, MAPK pathway, mouse embryonic fibroblasts, transcriptional activation
The tumor suppressor p53 plays a key role in regulating the cell cycle, apoptosis, genomic integrity, and DNA repair in response to various forms of genotoxic stress, and p53 is mutated or functionally impaired in most human cancers (1, 2). From the therapeutic point of view, it is important to devise strategies to induce apoptosis in the absence of functional p53, and a compound capable of doing so would be a good candidate for anticancer drug development. Earlier, we reported that green tea polyphenol, epigallocatechin-3-gallate (EGCG) differentially regulates the cellular growth of cancer cells in a p53-dependent manner through apoptosis and/or cell cycle arrest and established a crucial role of p53 and its two target proteins, p21 and Bax, in EGCG-induced apoptosis (3, 4).
In addition to p53, mammalian cells contain two closely related proteins, p63 and p73 (5, 6). Unlike p53, p63 and p73 mutations in human cancers are extremely rare (7). The p73 also has an alternative promoter within intron 3, from which a truncated p73 mRNA encoding truncated versions lacking the N-terminal transactivation domain (known as δNp73) is transcribed. Because the p73 protein functions as a tetramer, δNp73 acts as a dominant-negative suppressor of full-length p73 (8). The activity and protein stability of p73 is regulated by a number of complex posttranslational modifications that include ubiquitination, phosphorylation, prolyl-isomerization, recruitment into the PML-nuclear body (PML-NB), and acetylation (reviewed in refs. 9 and 10). Moreover, several proteins, such as Mdm2, Pin1, Notch, c-Myc, exportin-1, and many others directly interact with p73 and either increase or attenuate p73 transcriptional activity (reviewed in refs. 9 and 10).
In an attempt to further elucidate the pathways involved in differential negative growth regulation by EGCG, we explored the role of the tyrosine phosphatase SHP-2. Upon contact with many stimuli, SHP-2 is recruited to tyrosine-phosphorylated proteins and binds with various receptors and scaffolding adaptors (11–13). SHP-2 also regulates DNA damage-induced G2/M cell cycle arrest, most probably via Cdc2 phosphorylation, Cdc25C cytoplasmic translocation, and inactivation of p38 (14). A role of SHP-2 in cell survival has also been reported (15–17). In most receptor tyrosine kinase and cytokine signaling pathways, SHP-2 is required for full activation of the Erk/MAP cascade and for multiple receptor-evoked functions, including cell proliferation, differentiation, and migration (11, 12).
In this study, we find that SHP-2 protects cells from EGCG-induced apoptosis and that inactivation of SHP-2 renders the cells sensitive to EGCG. Moreover, EGCG-induced apoptosis is accompanied by the induction of a subset of p53 target genes, seemingly paradoxically, even in the absence of functional p53. We show that SHP-2 negatively regulates the expression of these genes and that the p53 family member p73 plays a critical role.
Results
SHP-2 Negatively Regulates Apoptosis Induced by EGCG.
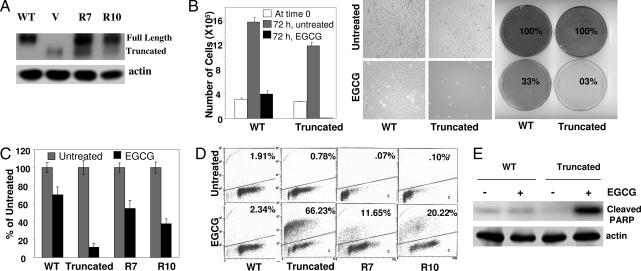
To investigate the mechanism of differential regulation of cell growth by EGCG, we used a pair of isogenic mouse embryonic fibroblasts (MEFs), expressing either WT or a functionally inactive/truncated SHP-2 (18). Because SHP-2 knockout mice die early in embryogenesis, MEFs were generated by immortalization with SV40 large T antigen, which renders p53 inactive. The expression of WT SHP-2 was restored in cells expressing inactive/truncated SHP-2 by introducing a plasmid containing WT SHP-2. Two independent clones were established by hygromycin selection (Fig. 1A). Cells were treated with 120 μM EGCG for 72 h. Live cells were counted by the tryphan blue dye exclusion method, plates were photographed and stained with methylene blue. As shown in Fig. 1B, there was a drastic reduction in the number of live cells after EGCG treatment in cells expressing inactive SHP-2. In contrast, there was a slight increase in the number of live cells after EGCG treatment in cells expressing WT SHP-2, indicating that cells continued growing for a while but eventually arrested. Methylene blue staining also provided similar results. We next tested the ability of the cells to resume normal growth in a colony-forming assay. Five hundred live cells after EGCG treatment were incubated in 15-cm plates in the absence of EGCG, and colonies were counted 3 weeks later. As shown in Fig. 1C, ≈70% of the WT cells resumed normal growth, whereas only 11% of cells expressing truncated SHP-2 formed colonies. The rescued clones also formed more colonies (54% and 37%). These results again suggest a pivotal role of SHP-2 in protecting cells from EGCG-induced cell death. To further investigate the mode of cell death, EGCG-treated cells were analyzed by Annexin V–propidium iodide staining using a kit from Calbiochem (La Jolla, CA) and the TUNEL assay using the APO-BRDU kit from Phoenix Flow Systems, which measures apoptosis by dual-color flow cytometry. As shown in Fig. 1D and supporting information (SI) Table 1, the great majority of cells expressing truncated SHP-2 stained positive in the TUNEL assay and Annexin V staining (66% and 48%, respectively). In contrast, the parental cells and the rescue clones expressing WT SHP-2 had much reduced TUNEL and Annexin V staining. As a molecular indicator of apoptosis, we also measured the degradation of PARP. As shown in Fig. 1E, PARP degradation was prevalent in cells expressing truncated SHP-2. All of the data together thus indicate that SHP-2 protects cells from EGCG-induced apoptosis.
Fig. 1.
SHP-2 negatively regulates EGCG-induced apoptosis. (A) Total cell lysates from cells expressing WT or truncated SHP-2 and two rescued clones (R7 and R10) were immunoblotted with anti-SHP-2. (B) Cells were treated with 120 μM EGCG for 72 h. Live cell numbers were counted by the trypan blue dye exclusion method, and plates were photographed and stained with methylene blue. (C) Cells were treated with EGCG for 72 h. Five hundred live cells were plated for colony formation. Results are plotted as percentages of untreated controls. (D) Cells were treated with EGCG for 72 h, fixed in ethanol, and stained with Apo-BrdU to determine the apoptotic population. (E) Cells were treated with EGCG for 24 h, and total cell lysates were immunoblotted with an antibody that specifically detects cleaved PARP (89 kDa) only.
To further investigate whether the catalytic activity of SHP-2 is important in protecting cells from EGCG-induced apoptosis, we used a cell line in which the catalytic activity of WT SHP-2 is disrupted by a dominant-negative SHP-2 (19). As shown in SI Fig. 6, the catalytic activity of SHP-2 is dispensable for its protective role from EGCG-induced apoptosis. However, inactivation of SHP-2 led to an increase in the G2/M population.
Regulation of p53 Target Genes by EGCG in Cells Undergoing Apoptosis.
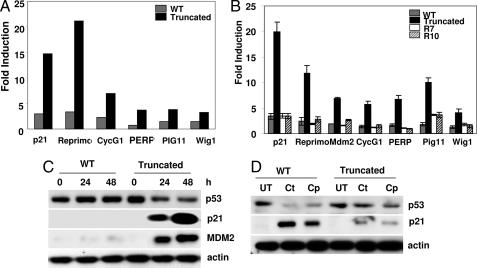
In an effort to further elucidate how SHP-2 protects cells from EGCG-induced apoptosis and affects the growth of these cells differentially, total RNAs from both cell types were analyzed after EGCG treatment using gene array technology. The array results are shown in SI Data Sets 1 and 2 and SI Fig. 7. A careful analysis of the data revealed that the expression of a number of genes involved in cell cycle regulation and apoptosis were differentially regulated by SHP-2 (Fig. 2A). Upon closer examination, we found that the great majority of these genes are known transcriptional targets of p53. The expression of these genes was greatly increased only in cells expressing an inactive SHP-2. In contrast, their expression was severely impaired in cells expressing WT SHP-2, suggesting that SHP-2 may act as a negative regulator for their expression. As shown in Fig. 2B, real-time PCR data completely corroborated the microarray results. Western blot analyses of a select number of proteins that are p53 transcriptional targets, such as Mdm2 and p21, further confirmed that these proteins are negatively regulated by SHP-2 (Fig. 2C). Because the response of these cells to EGCG was in sharp contrast to their response to DNA damage (20), we treated the cells with two DNA-damaging agents, camptothecin and cisplatin, and examined them for p21 expression. As shown in Fig. 2D, treatment with either camptothecin or cisplatin induced the expression of p21 in SHP-2 WT cells, and this expression was greatly impaired in cells with truncated SHP-2. Cell death was also apparent in cells with WT SHP-2 and not in those with truncated SHP-2 (data not shown). These experiments demonstrate that the effects of EGCG are different and distinct from the effects of activating the DNA-damage response pathway.
Fig. 2.
SHP-2 negatively regulates p53 target gene expression. (A) Cells were treated with 120 μM EGCG for 24 h. Total RNA was used for microarray analysis using the GE CodeLink mouse whole-genome chip containing 29,961 transcripts. Fold induction of p21, Reprimo, Cyclin G1 (CycG1), Perp, Pig11, and Wig1 are shown compared with an untreated control. (B) Cells were treated with 120 μΜ EGCG for 24 h. Total RNA was used in real-time PCR for the expression of p21, Reprimo, Mdm2, Cyclin G1, Perp, Pig11, and Wig1 mRNAs. (C) Cells were treated with 120 μM EGCG for the indicated times. Total cell lysates were immunoblotted with antibodies that detect p53, p21, or Mdm2. (D) Cells were treated with camptothecin (Ct) or cisplatin (Cp) for 24 h. Total cell lysates were immunoblotted with anti-p53 or anti-p21. UT, untreated.
SHP-2 Negatively Regulates Apoptosis and the Expression of p53 Target Genes in the Absence of p53.
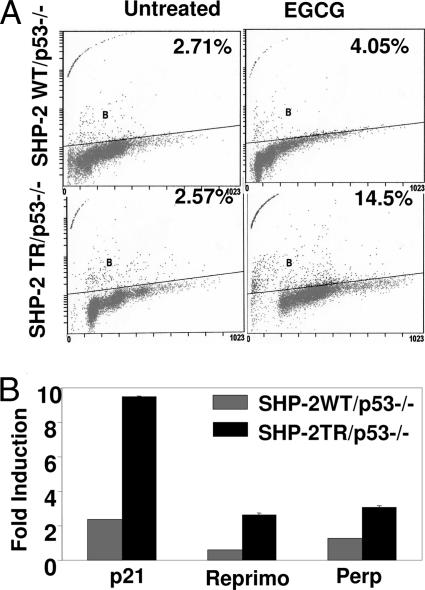
As shown above, the profile of expression of a set of genes that are p53 targets was modulated in cells that were immortalized by SV40 large T antigen, which inactivates p53. We next sought to unambiguously reveal whether the role of SHP-2 in EGCG-induced cellular responses depends on p53. For this purpose, we used an isogenic pair of cell lines (19). p53-null MEFs expressing either WT or truncated (functionally inactive) SHP-2 were treated with EGCG for 72 h, and the apoptotic population was determined by TUNEL staining. As shown in Fig. 3A, EGCG treatment led to pronounced apoptosis in cells expressing inactive SHP-2, whereas cells carrying WT SHP-2 were resistant. We next treated these cells with EGCG for 24 h and analyzed total RNA for the expression of p21, Reprimo, and Perp by real-time PCR. As shown in Fig. 3B, EGCG treatment induced the expression of p21, Reprimo, and Perp in cells with inactive SHP-2. In contrast, the expression of these genes was significantly suppressed in cells expressing WT SHP-2. These results further confirmed a negative regulatory role of SHP-2 in p53 target gene expression in the absence of p53. Because both of these cells were genetically p53-deficient, apoptosis and expression of p53 target gene are likely to be mediated by p53-independent signaling.
Fig. 3.
EGCG-induced apoptosis and p53 target gene expression in the absence of p53. (A) Cells genetically lacking p53 but expressing either WT or truncated SHP-2 were treated with 120 μM EGCG for 72 h. Cells were fixed in ethanol, and the apoptotic population was determined by TUNEL staining. (B) The same cells were treated with EGCG for 24 h. Total RNA was extracted and analyzed for the expression of p21, Reprimo, and Perp by real-time PCR.
Role of p73 in Regulation of p53 Target Genes by EGCG.
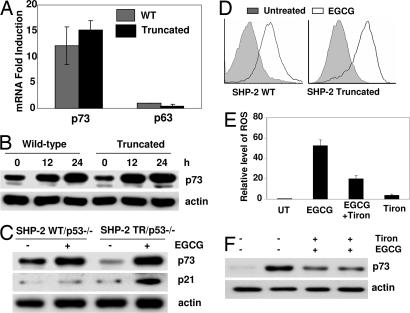
Because EGCG-induced apoptosis is accompanied by the induction of a number of p53 target genes in MEFs that were generated by immortalization with SV40 large T antigen, which selectively inactivates p53 but not p63 or p73 (21–25), we investigated the role and expression of these two additional members of the p53 family. Cells expressing either WT or truncated SHP-2 were treated with EGCG for 24 h, and total RNA was analyzed by real-time PCR. As shown in Fig. 4A, the expression of p63 mRNA remained unchanged in both cell types. In contrast, the expression of p73 mRNA in cells treated with EGCG was greatly increased, apparently to a comparable level in the two cell types. We next examined the expression of the p73 protein by Western blot analysis. Cells were treated with EGCG, and total cell lysates were immunoblotted with anti-p73. As shown in Fig. 4B, EGCG treatment resulted in increased expression of p73 protein in both cell lines. These observations taken together suggest that p73, but not p63, might be playing a role in the cellular response to EGCG.
Fig. 4.
Oxidative stress induced induction of p73 but not p63 by EGCG. (A) Cells were treated with EGCG for 24 h. Total RNA was analyzed for the expression of p73 and p63 by real-time PCR. (B) Cells were treated with EGCG for the indicated times. Total cell lysates were analyzed by Western blotting and probed with anti-p73. (C) Cells genetically lacking p53 but expressing either a WT or truncated SHP-2 were treated with EGCG for 24 h. Total cell lysates were immunoblotted with anti-p73 or anti-p21. (D) Cells were treated with EGCG for 24 h, and intracellular ROS generation was measured by HE staining. (E and F) SHP-2 mutant cells were treated with EGCG alone, EGCG plus 5 mM Tiron, or 5 mM Tiron alone for 24 h. Tiron was added 2 h before the addition of EGCG. ROS was measured by HE staining, and whole cell lysates were immunoblotted with anti-p73. UT, untreated.
To further investigate whether, in the absence of p53, p73 was responsible for the expression of p53 target genes, p53-null MEFs expressing either WT or truncated SHP-2 were treated with EGCG for 24 h, and total cell lysates were immunoblotted with anti-p73 or anti-p21. As shown in Fig. 4C, EGCG induced the expression of p73 protein in both cell types in a manner similar to that found in MEFs immortalized by SV40 large T antigen. These results suggest that, in the absence of p53, the family member p73 may be involved in regulating the expression of p53 targets such as p21. However, p21 protein expression was increased only in cells lacking functional SHP-2 and not in cells with WT SHP-2. This observation again confirmed that SHP-2 acts as a negative regulator of p53-target gene expression by EGCG.
Oxidative stress is one of the major signals that activate many apoptotic pathways. Although EGCG is known for its antioxidant properties, an increasing body of evidence suggests that EGCG also induces oxidative stress (26, 27). To identify the signal for p73 activation, we measured the generation of reactive oxygen species (ROS) in both SHP-2 WT and mutant cells. As shown in Fig. 4D, treatment with EGCG induced the generation of ROS in both the cell lines. To further investigate whether ROS was the signal for p73 induction, SHP-2 mutant cells were treated with a known antioxidant, Tiron. As shown in Fig. 4 E and F, pretreatment with Tiron inhibits the generation of ROS and also substantially inhibits EGCG-induced p73 induction. However, there is a small increase in p73 protein by Tiron itself, suggesting a ROS-independent regulation pf p73 induction. All these data suggest that EGCG treatment induces oxidative stress, which may activate p73.
Inactivation of p73 Reverses Apoptosis and Expression of p53 Target Genes.
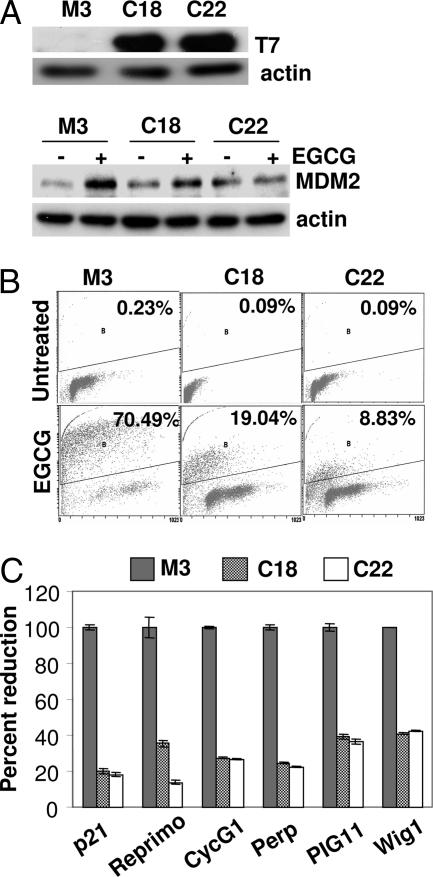
As described above, the treatment of EGCG results in comparable levels of p73 mRNA and protein induction in both SHP-2 WT and mutant cells. Because p73 activity is known to be modulated by a number of complex posttranslational events such as ubiquitination, phosphorylation, isomerization, acetylation, and interaction with several other proteins, we sought to further explore whether p73 is necessary for the regulation of p53 target genes and apoptosis by EGCG. p73 activity was disrupted by overexpressing dominant-negative p73 (DDp73) in cells expressing truncated SHP-2. The DDp73 mutant is tagged with a T7-epitope and has been shown to inactivate the activity of p73 (28). Two independent clones (C18 and C22) expressing DDp73 as determined with T7 antibody were established by G418 selection (Fig. 5A). Expression of Mdm2, a known target of p53, previously found to be induced by EGCG (Fig. 2) was also examined in these clones after EGCG treatment (Fig. 5A Lower). The EGCG-induced expression of Mdm2 is suppressed in these clones. The clones were treated with EGCG along with parental cells, and the apoptotic population was determined by TUNEL staining. As shown in Fig. 5B, inactivation of p73 substantially inhibits apoptosis in both the clones. We next treated these cells with EGCG for 24 h and analyzed total RNA for the expression of p21, Reprimo, Cyclin G1, Perp, Wig1, and Pig11. As shown in Fig. 5C, the expression of all of these genes was markedly inhibited after p73 inactivation. These observations together establish that, in the absence of functional p53, its family member p73 indeed regulates the expression of p53 target genes and apoptosis.
Fig. 5.
p73-dependent expression of p53 target genes and apoptosis in response to EGCG. (A) Cells expressing inactive SHP-2 (M3) were transfected with a T7-tagged dominant-negative p73 plasmid, and two clones (C18 and C22) were established. (Upper) Total cell lysates were probed with anti-T7. (Lower) Cells were treated with EGCG for 24 h, and total cell lysates were analyzed by the Western blot method, using anti-Mdm2. (B) Cells were treated with EGCG for 72 h, and the apoptotic population was measured by TUNEL staining. (C) Cells were treated with EGCG for 24 h, and total RNA was analyzed by real-time PCR for expression of the corresponding genes.
JNK Activation Is Dispensable for Apoptosis and p53 Target Gene Expression.
As mentioned earlier, SHP-2 is important for the regulation of p38 and JNK and is required for full activation of the Erk/MAP kinase pathway (11, 12). To further investigate the mechanism of differential role of SHP-2 in cellular growth regulation by EGCG, we examined the activation of JNK, Erk, p38, and Akt. As shown in SI Fig. 8A, EGCG treatment induced the phosphorylation of JNK in cells with truncated SHP-2, which was severely impaired in cells expressing WT SHP-2. In contrast, a transient phosphorylation of Erk was observed after 10 min in SHP-2 WT cells but not in SHP-2 mutant cells. Phosphorylation of Akt was markedly increased upon treatment with EGCG, and the level of induction was comparable between the two cell lines. However, there was no phosphorylation of p38 after EGCG treatment (data not shown). These results suggest that SHP-2 acts as a negative regulator for EGCG-induced JNK activation but as a positive regulator for Erk activation. These results also correlated with the known roles of these proteins in apoptosis. Because JNK activation regulates stress-induced apoptosis, its downstream target c-Jun is important for p73 activation (29), and a recent report suggests the involvement of JNK in EGCG-induced apoptosis (26), we next focused on JNK activation. To further confirm that SHP-2 negatively regulates EGCG-induced JNK phosphorylation, we analyzed JNK activation in the rescued clones. As shown in SI Fig. 8B, EGCG-induced JNK phosphorylation was suppressed after reintroduction of SHP-2. These results again suggest that SHP-2 is a negative regulator of EGCG-induced JNK phosphorylation. To test the importance of JNK activation in EGCG-induced p53 target gene expression, cells expressing truncated SHP-2 were pretreated with the specific JNK inhibitor, SP600125, for 1 h, followed by EGCG treatment for 24 h. Total RNA was analyzed for the expression of p21, Reprimo, Cyclin G1, Perp, Pig11, and Wig1 by real-time PCR. As shown in SI Fig. 8C, pretreatment with SP600125 had no significant effect on the expression of these genes. To further confirm that JNK activation is dispensable for EGCG-induced apoptosis, cells expressing truncated SHP-2 were pretreated with SP600125, followed by EGCG treatment for 72 h. As shown in SI Fig. 8D, inhibition of JNK activation had no significant effect on cell growth or apoptosis induced by EGCG.
No Direct Interaction of SHP2 with Tyrosine Phosphorylated Proteins or p73.
It is well established that SHP-2 becomes activated by binding of its SH2 domain to a phosphotyrosine residue of a target protein (11, 12). To investigate whether treatment with EGCG generates any tyrosine phosphorylated protein, total cell lysates from both the cell types were immunoblotted with an antiphosphotyrosine antibody. As shown in SI Fig. 9A, EGCG-treatment did not induce tyrosine phosphorylation of any cellular protein. Furthermore, SHP-2 immunoprecipitates from whole-cell lysates prepared with a buffer that does not interfere with complex formation (30) were immunoblotted with antiphosphotyrosine antibody to detect whether SHP-2 binds to any tyrosine phosphorylated protein. As shown in SI Fig. 9B, no additional bands were detected, thus suggesting that SHP-2 is not phosphorylated by EGCG nor does it interact with any tyrosine phosphorylated protein. We also investigated whether SHP-2 physically binds to p73. SHP-2 immunoprecipitates were immunoblotted with anti-p73. Again, as shown in SI Fig. 9C, no binding was detected.
Discussion
p53 plays a central role in the apoptotic response to chemotherapeutic agents. It was reported previously that EGCG had remarkable chemopreventive and chemotherapeutic properties, due mainly to its ability to induce apoptosis (3, 4, 31–33). In this study, we found that the protein tyrosine phosphatase SHP-2 serves as a negative regulator for EGCG-induced apoptosis because its inactivation renders cells sensitive to EGCG. However, the catalytic activity of SHP-2 is not important in mediating its protective role. SHP-2 is known to be involved in multiple signaling pathways that regulate cellular growth, survival, differentiation, apoptosis, and migration (11–14, 19, 20, 34). The roles of SHP-2 in mediating cellular growth and survival by activating Ras-MAPK and Akt signaling, thereby protecting cells from apoptosis, are well studied. In contrast, some studies suggest that SHP-2 is involved in mediating apoptosis in response to DNA damage or deprivation of growth factors (19, 20). However, the downstream targets of SHP-2 in mediating cell survival or apoptosis are not well characterized. Although earlier studies have identified SHP-2 as a positive regulator of apoptosis in response to DNA damage, our current observations suggest a completely opposite role of SHP-2 as a negative regulator of apoptosis in response to a well known chemopreventive agent in the very same cells. This intriguing observation further validates the notion that a single protein can influence signaling in diverse ways and thus influence the biological outcome.
Our observations establish that SHP-2 negatively regulates the expression of a subset of genes that are involved in cell cycle arrest and apoptosis and that have been well characterized as bona fide transcriptional targets of p53. Furthermore, the current findings also demonstrate that the expression of these genes is mediated in a p53-independent but p73-dependent manner because the MEFs used in the present study were either genetically p53-deficient or immortalized by SV40 large T antigen, which inactivates p53. Although some of these genes had been reported to be induced by p73, many were not. Our current observations also indicate that SHP-2 confers a protective function, probably by disabling p73 activity, thus inhibiting the expression of these genes.
Unlike p53, p73 and p63 are frequently functional and even overexpressed in some tumors (35). Therefore, these transcription factors are now considered to be an important determinant of cellular sensitivity to anticancer drugs, particularly in tumors lacking functional p53. In a search for the mechanism of apoptosis in the absence of p53, we found that EGCG treatment induced the expression of p73 but not p63 and that inactivation of p73 protects cells from EGCG-induced apoptosis. The results of our study are consistent with previous reports that p73 is sufficient to induce apoptosis in cells lacking functional p53 (36–38). However, there is no notable difference in the expression levels of p73 between SHP-2 WT and mutant cells, although there are remarkable differences in apoptosis and expression of target genes. We also did not find any differential distribution of p73 in the nuclear and cytosolic fractions or activation of p38, a kinase that activates p73 (data not shown). In addition to its subcellular localization, and explained earlier in the introduction section, the activity of p73 is modulated by a number of complex posttranslational events that include ubiquitination, sequential phosphorylation, prolyl-isomerization, recruitment into the PML-nuclear body (PML-NB), acetylation, and interaction with a large number of proteins. We believe that SHP-2 modulates one or more of these events to modulate p73 function. Obviously, a systematic study to identify as how SHP-2 regulates p73 activity is warranted.
The downstream targets required for p53 and its family members to execute apoptosis are many and vary with the cell type and stimulus. The BH3-only protein Bax, PUMA, and NOXA play important roles in mitochondrial apoptotic pathways induced in p53-dependent and -independent manners (3, 39–41). Perp, Pig11, and Wig1 are also involved in apoptosis (42–44). Another important p53 target p21 is well known for protecting cells in response to a variety of signals by halting the cell cycle at both the G1 and G2/M checkpoints (1, 2). We have also previously reported that both p21 and Bax play an important role in p53-dependent apoptosis induced by EGCG (3, 4). Although the roles of these p53 targets in apoptosis have been well studied, it is not yet clear which factors regulate their induction or lead to the expression of which targets. We found that treatment of cells with EGCG induced a number of these genes, including Cyclin G1, Perp, Wig1, and Pig11. A more in-depth study by ablating the expression of one protein at a time is needed to establish their role in apoptosis.
In conclusion, our study identifies SHP-2 as a protective factor of cells lacking functional p53 from EGCG-induced apoptosis and reveals a number of targets for EGCG-induced apoptosis that are expressed in the absence of functional p53 and that have not previously been reported to be so induced. Moreover, this report identifies a crucial role for p73 in EGCG-induced apoptosis and identifies a number of previously unidentified p73 target genes. Therefore, our study will hopefully open an avenue for apoptosis research, particularly in cells lacking functional p53, and will lead to the development of new anticancer drugs. A detailed mechanistic study is warranted to understand the negative regulatory role of SHP-2 in apoptosis and p53 target gene expression.
Materials and Methods
Cell Culture, Plasmid Transfection, and Cell Treatment.
All cells were maintained in DMEM containing 10% FBS. WT SHP-2 plasmid in pCDNA3-Hygro vector has been described earlier (18). Dominant-negative p73 (amino acids 327–636) plasmid was obtained from David Boothman (University of Texas Southwestern, Dallas, TX). Generation and validation of the plasmid is described elsewhere (28). SHP-2 mutant cells were transfected with these plasmids by using the Lipofectamine Plus reagent according to the supplier's instructions. Individual clones were selected by using hygromycin and G418. For EGCG treatment, 4 × 105 cells were plated in 10-cm culture dishes. After overnight incubation, the media were replaced, and the cells were treated with 120 μM EGCG for 72 h, if not indicated otherwise.
Materials and Chemicals.
EGCG was obtained from Tokyo Food Techno (Tokyo, Japan). The specific JNK inhibitor SP600125 was purchased from Calbiochem. Antibodies that recognize p42/44, p38 MAPK, JNK, Akt, SHP-2, Cleaved PARP, and Akt were from Cell Signaling Technology (Danvers, MA); Erk2, Bax, p21, p53, and SHP-2 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); T7-Tag antibody was from Novagen (San Diego, CA); and p73 antibody was from Bethyl Laboratories (Montgomery, TX).
TUNEL Assay.
After treatment with 120 μM EGCG for 72 h, cells were fixed in 70% ethanol and labeled with fluorescein-tagged bromodeoxy-uridine triphosphate (Brd-UTP) and propidium iodide as in the manufacturer's protocol, by using the APO-BRDU apoptosis kit (Phoenix Flow Systems, San Diego, CA).
Annexin V Staining.
Annexin V staining was performed by using a kit from Calbiochem according to the supplier's instruction. Briefly, after treatment with EGCG, cells were stained with Annexin V-FITC, followed by propidium iodide and were analyzed immediately by flow cytometry.
RNA Isolation and Microarray Analysis.
Total RNA was extracted from the cells by using the Qiagen RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Biotin-labeled cRNA was prepared by linear amplification of the Poly (A)+ RNA population within the total RNA sample. Purified cRNA was fragmented to uniform size and applied to CodeLink Bioarrays (GE Healthcare, Piscataway, NJ) in hybridization buffer. After hybridization, arrays were washed and stained with Cy5-Streptavidin dye conjugate. Dried arrays were scanned with a GenePix 4000B scanner. Arrays were processed with CodeLink Expression Analysis software (GE Healthcare), and data were analyzed with GeneSpring software (Silicon Genetics, Redwood City, CA). To compare individual expression values across arrays, raw intensity data (generated from CodeLink Expression 2 software) from each gene was normalized to the median intensity of the array. Only genes which have values greater than background intensity in at least one condition were used for further analysis. The arrays used in our experiment were GE CodeLink mouse whole-genome arrays. Information on these arrays can be seen at www5.amershambiosciences.com/aptrix/upp01077.nsf/Content/codelink_uni.
Measurement of ROS.
Intracellular ROS generation was measured by flow cytometry following staining with hydroethidium (HE), which is oxidized to ethidium bromide. Briefly, after treatment with EGCG for 24 h, cells were stained with HE and analyzed by using an Epics XL Flow Cytometer (Coulter, Miami, FL).
Real-Time PCR.
Total RNA extracted as above was used for real-time PCR. Amplification of the corresponding genes was done by using primer sets supplied by Applied Biosystems (Foster City, CA). The data were analyzed for fold induction of each gene as compared with the untreated sample.
Western Blot Analyses.
Total cellular proteins were isolated by lysing the cells in 20 mM Tris·HCl, pH 7.5, 2% (wt/vol) SDS, 2 mM benzamidine, and 0.2 mM phenylmethanesulfonyl fluoride. Protein concentrations were determined by the Bradford method (Bio-Rad, Hercules, CA). Proteins were resolved on SDS-10% polyacrylamide gels and then transferred to polyvinylidene diflouride (PVDF) membranes. The membranes were blocked in 5% nonfat skimmed milk and incubated with the antibody, followed by incubation with a secondary antibody. Proteins were visualized by using Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Boston, MA) as directed by the manufacturer.
Supplementary Material
Acknowledgments
We thank David Boothman for the dominant-negative p73 construct and GenUs Biosystems, Inc. (Northbrook, IL) for conducting the micorarray analysis. This work was supported by National Institutes of Health Grants R01 CA98916 (to M.L.A.) and CA78809, CA101039, and DK065303 (to H.M.).
Abbreviations
- EGCG
epigallocatechin-3-gallate
- HE
hydroethidium
- MEF
mouse embryonic fibroblast
- ROS
reactive oxygen species.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700642104/DC1.
References
- 1.Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Hastak K, Agarwal MK, Mukhtar H, Agarwal ML. FASEB J. 2005;19:789–791. doi: 10.1096/fj.04-2226fje. [DOI] [PubMed] [Google Scholar]
- 4.Hastak K, Gupta S, Ahmad N, Agarwal MK, Agarwal ML, Mukhtar H. Oncogene. 2003;22:4851–4859. doi: 10.1038/sj.onc.1206708. [DOI] [PubMed] [Google Scholar]
- 5.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J, Valent A, Minty A, Chalon P, Lelias J, Dumont X, et al. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 6.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 7.Vousden KH, Lu X. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 8.Grob TJ, Novak U, Maisse C, Barcaroli D, Luthi AU, Pirnia F, Hugli B, Graber HU, De Laurenzi V, Fey MF, et al. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- 9.Melino G, Lu X, Gasco M, Crook T, Knight RA. Trends Biochem Sci. 2003;28:663–670. doi: 10.1016/j.tibs.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Oberst A, Rossi M, Salomoni P, Pandolfi PP, Oren M, Melino G, Bernassola F. Biochem Biophys Res Commun. 2005;331:707–712. doi: 10.1016/j.bbrc.2005.03.158. [DOI] [PubMed] [Google Scholar]
- 11.Neel BG, Gu H, Pao L. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 12.Feng GS. Exp Cell Res. 1999;253:47–54. doi: 10.1006/excr.1999.4668. [DOI] [PubMed] [Google Scholar]
- 13.Amin ARMR, Machida K, Oshima K, Oo ML, Thant AA, Senga T, Matsuda S, Akhand AA, Maeda A, Kurosaki T, et al. Oncogene. 2002;21:8871–8878. doi: 10.1038/sj.onc.1206018. [DOI] [PubMed] [Google Scholar]
- 14.Yuan L, Yu WM, Qu CK. J Biol Chem. 2003;278:42812–82820. doi: 10.1074/jbc.M305075200. [DOI] [PubMed] [Google Scholar]
- 15.Zhan Y, O'Rourke DM. Cancer Res. 2004;64:8292–8298. doi: 10.1158/0008-5472.CAN-03-3143. [DOI] [PubMed] [Google Scholar]
- 16.Ivins Zito C, Kontaridis MI, Fornaro M, Feng GS, Bennett AM. J Cell Physiol. 2004;199:227–236. doi: 10.1002/jcp.10446. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Yu WM, Bunting KD, Qu CK. Oncogene. 23:3659–3669. doi: 10.1038/sj.onc.1207471. [DOI] [PubMed] [Google Scholar]
- 18.Shi ZQ, Lu W, Feng GS. J Biol Chem. 1998;273:4904–4908. doi: 10.1074/jbc.273.9.4904. [DOI] [PubMed] [Google Scholar]
- 19.Yuan L, Yu WM, Xu M, Qu CK. J Biol Chem. 2005;280:42701–42706. doi: 10.1074/jbc.M506768200. [DOI] [PubMed] [Google Scholar]
- 20.Yuan L, Yu WM, Yuan Z, Haudenschild CC, Qu CK. J Biol Chem. 2003;278:15208–15216. doi: 10.1074/jbc.M211327200. [DOI] [PubMed] [Google Scholar]
- 21.Marin MC, Jost CA, Irwin MS, DeCaprio JA, Caput D, Kaelin WG., Jr Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheppard HM, Corneillie SI, Espiritu C, Gatti A, Liu X. Mol Cell Biol. 1999;19:2746–2753. doi: 10.1128/mcb.19.4.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobbelstein M, Roth J. J Gen Virol. 1998;79:3079–3083. doi: 10.1099/0022-1317-79-12-3079. [DOI] [PubMed] [Google Scholar]
- 24.Kojima T, Ikawa Y, Katoh I. Biochem Biophys Res Commun. 2001;281:1170–1175. doi: 10.1006/bbrc.2001.4486. [DOI] [PubMed] [Google Scholar]
- 25.Roth J, Dobbelstein M. J Gen Virol. 1999;80:3251–3255. doi: 10.1099/0022-1317-80-12-3251. [DOI] [PubMed] [Google Scholar]
- 26.Qanungo S, Das M, Haldar S, Basu A. Carcinogenesis. 2005;26:958–967. doi: 10.1093/carcin/bgi040. [DOI] [PubMed] [Google Scholar]
- 27.Elbling L, Weiss RM, Teufelhofer O, Uhl M, Knasmueller S, Schulte-Hermann R, Berger W, Micksche M. FASEB J. 2005;19:807–809. doi: 10.1096/fj.04-2915fje. [DOI] [PubMed] [Google Scholar]
- 28.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, Kaelin WG., Jr Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 29.Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. J Biol Chem. 2004;279:44713–44722. doi: 10.1074/jbc.M407672200. [DOI] [PubMed] [Google Scholar]
- 30.Wong BR, Besser D, Kim N, Arron JR, Vologodskaia M, Hanafusa H, Choi Y. Mol Cell. 1999;4:1041–1049. doi: 10.1016/s1097-2765(00)80232-4. [DOI] [PubMed] [Google Scholar]
- 31.Zaveri NT. Life Sci. 2006;78:2073–2080. doi: 10.1016/j.lfs.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Moyers SB, Kumar NB. Nutr Rev. 2004;62:204–211. doi: 10.1111/j.1753-4887.2004.tb00041.x. [DOI] [PubMed] [Google Scholar]
- 33.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 34.Yu DH, Qu CK, Henegariu O, Lu X, Feng GS. J Biol Chem. 1998;273:21125–21131. doi: 10.1074/jbc.273.33.21125. [DOI] [PubMed] [Google Scholar]
- 35.Zaika AI, Kovalev S, Marchenko ND, Moll UM. Cancer Res. 1999;59:3257–3263. [PubMed] [Google Scholar]
- 36.Tomasini R, Seux M, Nowak J, Bontemps C, Carrier A, Dagorn JC, Pebusque MJ, Iovanna JL, Dusetti NJ. Oncogene. 2005;24:8093–8104. doi: 10.1038/sj.onc.1208951. [DOI] [PubMed] [Google Scholar]
- 37.Allart S, Martin H, Detraves C, Terrasson J, Caput D, Davrinche C. J Biol Chem. 2002;277:29063–29068. doi: 10.1074/jbc.M201974200. [DOI] [PubMed] [Google Scholar]
- 38.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG., Jr Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 39.Shibue T, Suzuki S, Okamoto H, Yoshida H, Ohba Y, Takaoka A, Taniguchi T. EMBO J. 2006;25:4952–4962. doi: 10.1038/sj.emboj.7601359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karst AM, Dai DL, Cheng JQ, Li G. Cancer Res. 2006;66:9221–9226. doi: 10.1158/0008-5472.CAN-05-3633. [DOI] [PubMed] [Google Scholar]
- 41.Obexer P, Geiger K, Ambros PF, Meister B, Ausserlechner MJ. Cell Death Differ. 2007;14:534–547. doi: 10.1038/sj.cdd.4402017. [DOI] [PubMed] [Google Scholar]
- 42.Ihrie RA, Reczek E, Horner JS, Khachatrian L, Sage J, Jacks T, Attardi LD. Curr Biol. 2003;13:1985–1990. doi: 10.1016/j.cub.2003.10.055. [DOI] [PubMed] [Google Scholar]
- 43.Liang XQ, Cao EH, Zhang Y, Qin JF. Oncol Rep. 2003;10:1265–1269. [PubMed] [Google Scholar]
- 44.Varmeh-Ziaie S, Okan I, Wang Y, Magnusson KP, Warthoe P, Strauss M, Wiman KG. Oncogene. 1997;15:2699–2704. doi: 10.1038/sj.onc.1201454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.