Abstract
PTEN is an important tumor-suppressor gene associated with many cancers. Through expression profiling of glioblastoma tissue samples and prostate cancer xenografts, we identified a molecular signature for loss of the PTEN tumor suppressor in glioblastoma and prostate tumors. The PTEN signature consists of a minimum of nine genes, several of which are involved in various pathways already implicated in tumor formation. Among these signature genes, the most significant was an increase in insulin growth factor-binding protein 2 (IGFBP-2) mRNA. Up-regulation of IGFBP-2 was confirmed at the protein level by Western blot analysis and validated in samples not included in the microarray analysis. The link between IGFBP-2 and PTEN was of particular interest because elevated serum IGFBP-2 levels have been reported in patients with prostate and brain tumors. To further investigate this link, we determined that IGFBP-2 expression is negatively regulated by PTEN and positively regulated by phosphatidylinositol 3-kinase (PI3K) and Akt activation. In addition, Akt-driven transformation is impaired in IGFBP2−/− mouse embryo fibroblasts, implicating a functional role for IGFBP-2 in PTEN signaling. Collectively, these studies establish that PTEN and IGFBP-2 expression are inversely correlated in human brain and prostate cancers and implicate serum IGFBP-2 levels as a potential serum biomarker of PTEN status and PI3K Akt pathway activation in cancer patients.
Keywords: brain cancer, microarray
Using the tools of global expression profiling, it has become possible to classify many tumors from the same site of origin into distinct clinical subgroups, to implicate genes involved in tumor formation and progression and to define patients with distinct prognoses (1–6). Expression profiling can also define signatures of specific signaling pathways, such as those of Myc, Ras, and other oncogenes (7, 8). Moreover, gene expression signatures have been mined to yield functional linkages, such as the role of C/EBPβ/Nf-Il6 transcription factor in cyclin D1 overexpression (9).
One challenge in the clinical development of molecularly targeted cancer therapy is the selection of appropriate patients, i.e., those whose tumors reflect alterations in the specific pathways to be targeted. The feasibility of conducting large-scale clinical trials in such molecularly defined populations would be greatly facilitated by the availability of serum biomarkers that might reflect the activation state of a signaling pathway in the tumor. Prostate-specific antigen is one example of such a pathway biomarker, because it reflects activation of the androgen receptor and serves as an excellent readout for the efficacy of antiandrogen therapy in men with hormone-refractory prostate cancer (10). Here we have used global expression profiling in a cohort of human glioblastoma and prostate samples annotated for PTEN status to define a molecular signature that might include biomarkers with clinical utility.
PTEN is a tumor-suppressor gene implicated in multiple human cancers through mutation, homozygous deletion, loss of heterozygosity, or epigenetic loss of expression (11–13). Among the most cancers most commonly affected by PTEN abnormalities are glioblastoma, prostate, and endometrial tumors, with frequencies approaching 50% (14). Germline PTEN mutations are found in two autosomal-dominant cancer predisposition syndromes (14, 15). Homozygous deletion of the PTEN gene in mice is lethal, and heterozygous deletion results in tumor formation in several organs (16–18). The best characterized biochemical function for the PTEN protein is its lipid phosphatase activity against phosphatidylinositol triphosphate (PIP3), the product of phosphoinositide 3-kinase (PI3K) activation in response to many growth factors implicated in tumor formation (18, 19). This PI3K-antagonizing activity in turn inhibits activation of the downstream effector Akt and leads to inhibition of cell survival and proliferation, cellular processes essential for tumor formation and progression (20).
To identify transcriptional targets associated with PTEN loss, we performed microarray-based expression profiling with on a panel of human prostate cancer xenografts and human glioblastoma samples annotated for PTEN status by mutation analysis and protein expression. Because signaling networks are system- and context-dependent, we felt that it was important not to rely strictly on cell lines. Therefore we included xenografts, which consist of nearly homogeneous human tumor material, and archived clinical material selected for high human tumor cell content. Through this analysis we identified a nine-gene signature that accurately classified the tumor samples according to PTEN status regardless of tumor type. We also show that the most significant gene in the signature, insulin growth factor-binding protein 2 (IGFBP-2), is inversely regulated by PTEN in isogenic models and required for Akt-mediated fibroblast transformation. Interestingly, elevated serum IGFBP-2 levels have been reported in patients with prostate cancer and glioblastoma, as well as other cancers such as colorectal, ovarian and acute lymphoblastic leukaemia (21–24), but not previously linked to PTEN status. Therefore, our findings raise the possibility that serum levels of IFGBP-2, likely in conjunction with the measurement of other markers, could serve as a biomarker of PTEN status in patients with prostate cancer, glioblastoma or perhaps other cancers with abnormalities in the PI3K/PTEN/Akt pathway.
Results
A Nine-Gene mRNA Classifier for Loss of the PTEN Tumor-Suppressor Gene.
To identify transcriptional targets associated with PTEN tumor-suppressor function, we compared the gene expression profiles of 11 tissue or xenograft samples that bear the wild-type PTEN gene to those of 14 samples with loss of PTEN function as defined by deletion, mutation, or loss of protein expression (Fig. 1). These 25 samples included 12 advanced prostate cancer xenografts (reflecting six isogenic hormone-sensitive and hormone-refractory sublines) and 13 glioblastoma tissue samples. The PTEN status of the prostate cancer xenografts were characterized in ref. 25. The glioblastoma samples were examined by Western blot analysis and genomic DNA sequence analysis. Six samples had no detectable PTEN protein expression and were classified as PTEN mutant. The other seven samples were classified as PTEN wild type on the basis of easily detected PTEN protein expression and the absence of point mutations, as determined by genomic DNA sequence analysis (data not shown).
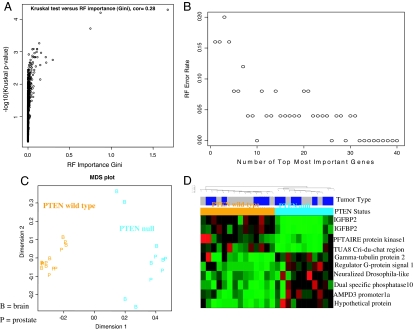
Fig. 1.
Identification of a gene expression classifier of PTEN status. Microarrays were used to measure the expression profiles of 25 prostate cancer xenograft and glioblastoma samples, of which 11 were classified PTEN wild type and 14 were classified PTEN mutant. To relate PTEN status to the gene expression data, we used RF predictors and the Kruskal–Wallis test of differential expression. An RF importance measure was used to rank probe sets according to their prognostic importance for PTEN status. (A) The RF variable importance measure is highly correlated (r = 0.28) with a conventional measure of differential expression (minus log 10 of the Kruskal–Wallis test P value). In the SI Appendix, we also consider alternative variable importance measures and the Student t test. We find that our results are highly robust with respect to the gene screening method. (B) The error rate of the RF predictor as a function of the number of most important probe sets. We find that the 10 most important probe sets lead to an apparent error rate of zero. (C) Classical multidimensional scaling plot for visualizing the dissimilarities between the microarray samples (on the basis of the 10 most important probe sets). Prostate cancer samples are labeled “P,” and glioblastoma samples are labeled “B.” PTEN mutant samples are in orange, and wild type is in cyan (turquoise). The plot in C and the supervised hierarchical clustering plot (D) show that the 10 most important probe sets stratify the samples according to their PTEN status.
The expression profiles of all 25 prostate cancer and glioblastoma samples were analyzed by using a random forest (RF) prediction method to identify a tissue-independent signature that might classify the tumors by PTEN status (26). As byproducts of its construction, an RF predictor yields a measure of variable importance which can be used to determine which gene expression values are most important for predicting the PTEN status (Fig. 1A). The larger the importance measure, then the more important the gene (or probe set) is for predicting PTEN status. The results obtained through RF analysis were consistent with those obtained by using other methods such as the Kruskal–Wallis test of differential expression (Fig. 1A) and the Student t test [supporting information (SI) Appendix]. The apparent error rate of the RF predictor can be measured as a function of the number of probe sets used to make the class distinction. For the 10 most important probe sets, the apparent error rate of the RF fell to zero (Fig. 1B). IGFBP-2 is represented twice in this top-10 list (by two independent probe sets), thus the classifier contains only nine genes. As an alternative gene-filtering method, we used the Kruskal–Wallis test, a nonparametric multigroup comparison test, to determine genes that are differentially expressed between wild-type and mutant PTEN samples. These same 10 probe sets are significantly differentially expressed (P < 0.0075) and are among the 30 most differentially expressed probe sets (SI Appendix and Tables 1 and 2) (only 9 of 10 of these probe sets are among the top 30 on the basis of Kruskal–Wallis test). The ability of the 10 probes to classify tumor samples with different PTEN status can also be visualized by a multidimensional scaling plot (Fig. 1C) and by supervised hierarchical clustering (Fig. 1D). Thus, we find that these nine genes can separate PTEN mutant from PTEN wild-type tumors in this data set regardless of where the cancer originated (glioblastoma or prostate cancer).
Elevated Levels of IGFBP-2 Protein Expression in PTEN Mutant Tumors.
We next examined the link between PTEN loss and up-regulation of IGFBP-2, the most significant gene in the classifier, at the protein level. Using lysates available from 10 prostate cancer xenografts and 24 glioblastoma tissue samples, we found increased levels of IGFBP-2 protein in all PTEN-null tumors. This analysis included four prostate xenografts and 10 glioblastomas that were not part of the original test set from which the nine-gene signature was derived (Fig. 2 A and B and SI Table 3). Glioblastoma sample 429 was notable because a high level of IGFBP-2 protein expression was observed despite wild-type PTEN status. Western blot analysis indicated high levels of Akt activation as measured by using the phospho-specific pSer473 antibody (data not shown), suggesting that Akt is activated by a mechanism other than PTEN mutation in this sample. This finding also raises the possibility that Akt activation is responsible for elevated expression of IGFBP-2 in PTEN-null cancers.
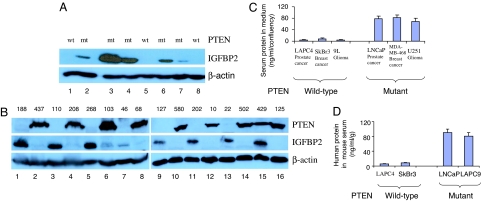
Fig. 2.
PTEN-null cancers express high levels of the secreted protein IGFBP-2. (A and B) IGFBP2 and PTEN protein levels are inversely correlated by Western blot analysis in previously characterized prostate cancer xenografts with known PTEN status LAPC4 (1), LAPC9 (2), LuCaP 35 (3), LAPC3 (4), LuCaP23 (5), LAPC12 (6), LuCaP41 (7), and LAPC14 (8) (A) and in glioblastoma tissue samples (B). (C and D) Human IGFBP2 levels were measured by RIA in the media of brain, prostate, and breast cancer cell lines (n = 2) (C) and in the sera of mice bearing human prostate and breast cancer xenografts mice (n = 2) (D). Values were normalized to cell confluency in B and to the weight of the excised xenograft tumor in C.
Because IGFBP-2 is a secreted protein, we asked whether elevated levels are detected in the supernatants of PTEN-null cell lines grown in culture. In a panel of six cell lines, including two each from prostate, glioblastoma, and breast cancer, IGFBP-2 levels measured by RIA were higher in the three PTEN-null lines (>70 ng/ml) than in the three PTEN wild-type lines (<20 ng/ml) when normalized for confluency (Fig. 2C). We extended these findings to xenograft models of prostate and breast cancer, detecting elevated serum levels of human IGFBP-2 in mice bearing s.c. PTEN-null tumors, but not PTEN wild-type tumors (Fig. 2D). Therefore, human tumors with PTEN mutations (including breast cancer) secreted high levels of IGFBP-2, which might serve as a serum biomarker for PTEN status.
Inhibition of IGFBP-2 Expression by PTEN.
To examine the molecular basis for the PTEN/IGFBP2 link, we extended our analysis to isogenic mouse embryonic fibroblasts (MEFs) isolated from PTEN knockout mice (27). Consistent with the human tumor findings, IGFBP-2 protein expression was substantially higher in PTEN−/− MEFs (Fig. 3A). To determine whether up-regulation of IGFBP-2 expression in PTEN-null cells depends on the PI3K/Akt pathway, we treated PTEN mutant LnCaP prostate cancer cells with the PI3K inhibitor LY294002. IGFBP-2 levels fell to baseline in drug-treated cells in parallel with reduced levels of Akt phosphorylation (Fig. 3B). Because LY294002 can inhibit other targets in addition to PI3K, we also pursued a genetic approach to address the PTEN/IGFBP-2 link. Whereas LY294002 lowered IGFBP-2 levels in PTEN-null cells, increased levels of IGFBP-2 expression were observed in PTEN wild-type DU145 prostate cancer cells infected with retrovirus expressing a constitutively active Akt allele (HA–Myr–Akt) (Fig. 3C). These results indicate that IGFBP-2 expression is induced by PI3K/Akt pathway activation and antagonized by the PTEN tumor suppressor.
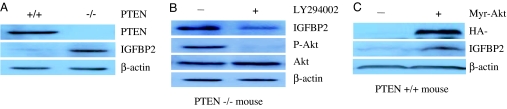
Fig. 3.
IGFBP2 expression is negatively regulated by PTEN and positively regulated by the PI3K/Akt pathway. (A) The levels of PTEN, IGFBP-2, and β-actin protein were measured in PTEN+/+ and PTEN−/− MEFs by Western blot analysis. (B) PTEN mutant LnCaP prostate cancer cells were treated with the PI3K inhibitor LY294002 and then were examined for IGFBP-2 protein expression and Akt activation by Western blot analysis. (C) PTEN wild-type DU145 prostate cancer cells were infected with retrovirus-expressing HA-tagged Myr–Akt and then examined for expression of the Akt transgene, IGFBP-2, and β-actin.
Functional Role of IGFBP-2 in PTEN/Akt Signaling.
IGFBP-2 functions as an insulin-like growth factor 1 (IGF1) binding protein and, in certain contexts, enhances the agonist effects of IGF1 on the IGF1 receptor (IGF1R) (28). Because IGF1R potently activates the PI3K/Akt pathway, one could envision a model in which the increased levels of IGFBP-2 observed in PTEN-null cells might play a functional role, perhaps by enhancing positive input to PI3K. To explore a potential functional link between IGFBP-2 and PTEN, we asked whether overexpression of IGFBP-2 in PTEN-null cells treated with a PI3K inhibitor might rescue any of the biological effects of PI3K inhibition. As expected, LY294002 treatment reduced the fraction of PTEN-null LnCaP prostate cancer cells in S phase and lowered IGFBP-2 levels. This reduction in S phase was partially rescued by the forced expression of exogenous IGFBP2, whereas IGFBP-2 expression had no measurable effect in the absence of LY294002 (Fig. 4A). We further addressed the functional role of IGFBP-2 by using IGFBP-2−/− MEFs. Infection of IGFBP-2+/+ MEFs with HA–Myr–Akt retrovirus led to focus formation when plated on plastic at limiting dilution and quantified by crystal violet visualization. The number of foci was reduced substantially in IGFBP-2−/− MEFs despite comparable levels of HA–Myr–Akt expression (Fig. 4B). This failure of constitutively active Akt to promote focus formation in IGFBP-2−/− MEFs was partially rescued by reintroduction of IGFBP-2 (Fig. 4B). Furthermore, the requirement of IGFBP-2 was specific to Akt because focus formation was observed in IGFBP-2+/+ and IGFBP-2−/− MEFs at comparable levels after infection with c-Myc retrovirus (Fig. 4C). These data suggest that increased IGFBP-2 levels, in addition to serving as a potential biomarker of PTEN loss, may also play a functional role. The mechanism by which IGFBP-2 might promote PI3K/Akt pathway transformation requires much further work. Although one possibility is a feed-forward loop through enhanced IGF1R activation, IGFBP-2 can also regulate the expression of genes involved in tumor invasion (29).
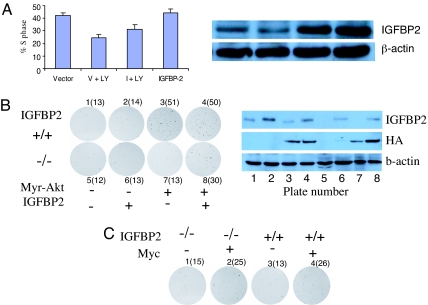
Fig. 4.
IGFBP2 plays a functional role in PTEN/Akt signal transduction. (A) LnCaP cells were treated with the PI3K inhibitor LY294002. IGFBP-2 expression was measured by Western blot analysis, and the percent of cells in S phase was determined as described in Methods. LY, LY294002; V, empty vector; I, IGFBP-2 cDNA. (B) IGFBP2+/+ (plates 1–4) or IGFBP2−/− (plates 5–8) MEFs infected with retrovirus expressing HA-epitope tagged Mry–Akt (plates 3, 4, 7, and 8), IGFBP2 (plates 2, 4, 6, and 8), or corresponding control vectors and then plated at limiting dilution on plastic as described in Methods. Colonies were visualized by crystal violet staining and counted after 2 weeks of culture. The results for Myr–Akt and Myc are quantified in B and C, respectively.
Discussion
We report here that nine genes serve as a classifier of PTEN status in a defined set of human glioblastoma samples and human prostate cancer xenografts. The most significant among them is the secreted protein IGFBP-2 (P = 7.5 × 10−7), which has been independently shown to be up-regulated in glioblastoma and prostate tumors with poor prognosis but not previously linked to PTEN status (30, 31).
Other Genes in the PTEN Classifier.
Although the focus of this report is on IGFBP-2, it is worth noting that several of the other eight genes in the PTEN classifier also have plausible biological roles in PI3K/Akt pathway function and may be worthy of further investigation. For example, Neuralized Drosophila-like is an E3 ligase for the Notch signal transduction pathway and mediates proteosome-dependent degradation of the Notch ligand Delta (32). Therefore, reduced levels of Neuralized, as seen in PTEN-null tumors, would be expected to lead to enhanced Notch activation, which is associated with many cancer phenotypes (33, 34). Indeed, loss-of-function mutations of the neurogenic genes produce hyperplasia of the embryonic nervous system in flies (35), somewhat analogous to the brain phenotype of the brain-specific PTEN knockout mice (36). Furthermore, Neuralized is highly expressed in normal human brain tissue but is low or absent in advanced gliomas (37).
Another classifier gene of interest is dual-specificity phosphatase 10 (also called MAPK phosphatase-5 or MKP-5), which selectively dephosphorylates JNK and reduces its activity (38). The microarray data show that DSP-10/MKP-5 levels are reduced in PTEN-null tumors, suggesting that JNK activity may be increased in PTEN-null cancers. A third gene in the classifier, Regulator of G protein signaling 1, is noteworthy because it specifically binds to and is inhibited by the PTEN substrate phosphatidylinositol triphosphate (PIP3) (39, 40), raising the possibility of feedback control of PI3K signaling.
IGFBP-2 as a Potential Biomarker of PTEN Status.
Our identification of IGFBP-2 up-regulation in PTEN-null glioblastomas and prostate cancers is consistent with prior reports examining IGFBP-2 levels in tumor samples and in patient sera (41). For example, IGFBP-2 overexpression was observed in up to 50% of glioblastomas but not in lower-grade astrocytomas and was associated with a worse prognosis (42, 43). The frequency and prognostic impact of IGFBP-2 overexpression in glioblastoma are remarkably similar to similar statistics on PTEN loss (44), consistent with the notion raised here that these events are linked. Indeed, while this work was underway, another group reported that IGFBP-2 protein level is regulated by PTEN in glioblastoma cells but did not define the mechanism (45). Further studies are required that use larger, independent data sets annotated for PTEN status to determine whether the nine-gene classifier and particularly IGFBP-2 serum levels can serve as clinical biomarkers of PTEN status.
Potential Utility of Pathway Biomarkers in Clinical Trials.
A large number of signaling pathway inhibitors are currently entering the clinic and are anticipated to have activity in subsets of patients, primarily those with abnormalities in the molecularly targeted pathway of interest. On the basis of the frequency of PTEN and PI3K pathway mutations in a broad range of tumor types, inhibitors of this pathway have been an area of intense interest. However, successful clinical development requires tools that permit classification of patients on the basis of these molecular lesions. Tumor genotyping will undoubtedly figure prominently in solving this problem, but companion serum markers that reflect the activation state of a particular signaling pathway could prove invaluable in classifying patients and in monitoring response to pathway-specific therapy. IGFBP-2 serum levels may be such a tool to monitor response to PI3K pathway inhibitors, analogous to the effects we observed here in PTEN-null model systems. Of note, others have observed that Hsp90 inhibitors, which promote Akt protein degradation (among other effects), lower IGFBP-2 serum levels in xenograft models (46). Conversely, elevated IGFBP-2 serum levels have been implicated as a biomarker for treatment with recombinant IGF1, which activates IGF1R and the PI3K/Akt pathway (47). In summary, we report here that PTEN status is inversely correlated with IGFBP-2 expression in prostate cancer xenografts and clinical glioblastoma samples and propose that serum IGFBP-2 levels may be a biomarker for PI3K/Akt pathway activation in patients with PTEN-null cancers.
Methods
Prostate Cancer Xenografts.
All of the prostate cancer xenografts (LAPC9, LUCaP 35, LAPC4, LUCaP23, LAPC3, LAPC12, LUCaP41, and LAPC14) were derived from patients that had local advanced or metastatic prostate cancer (48). The human specimens were implanted into the flanks of severe combined immunodeficient or nude mice in serial passages to enrich for homogenous populations of prostate cancer cells. Microarray expression profiling data from some of these xenografts were reported in a study of mechanisms of hormone refractory growth (49). Animals were maintained in accordance with institutional guidelines. The PTEN status of the prostate cancer xenografts was characterized in ref. 25.
Glioblastoma Samples.
To exclude any potential effect of treatment on gene expression, we studied glioblastoma multiforme (GBM) from untreated patients that had developed de novo (primary GBM) and from which high-quality RNA could be obtained. All samples were collected at the University of California, Los Angeles with informed consent.
Microarray Experiments.
For the glioblastoma samples, Affymetrix (Santa Clara, CA) U95av2 was used to interrogate 12,533 probe sets encoding ≈10,000 genes. For the prostate cancer xenografts, U95A arrays were used (Affymetrix). A list of common genes was constructed to compare expression in all samples. Hybridization to the genome chips were performed according to standard Affymetrix protocols. Briefly, 15 mg of total RNA was converted into double-stranded cDNA by using the SuperScript Choice System for first- and second-strand cDNA synthesis (GIBCO/BRL, Carlsbad, CA). T7 RNA polymerase mediated in vitro transcription reactions was used to convert double-stranded cDNA into the biotinylated single-stranded cRNA (Enzo BioArray High-Yield RNA transcript labeling kit; Enzo Diagnostics, Farmingdale, NY). The cRNA was randomly fragmented by chemical hydrolysis. ≈15 μg of cRNA target was applied to the Affymetrix chips for a 16-h hybridization (Oven 640). Using the Affymetrix GeneChip Fluidics Station 400, the unbound fragments were washed and the specific bound fragments were stained with a phycoerythrin–streptavidin conjugate that binds to the biotin label. The fluorescent signal for each chip was captured with a GeneChip Scanner. Photomultiplier output signal was quantified and converted into a digitized fluorescence image, and a single fluorescent intensity value was assigned to each probe.
Microarray Data Analysis.
Additional materials and methods as well as the statistical software code used for the RF analysis are provided in the SI Appendix. The normalized gene expression data and the corresponding gene information are included as SI Tables 1 and 2. The SI Appendix also serves as a tutorial for RF analysis with the R software. The .CEL files for all of the microarray hybridizations (27 samples) generated by Affymetrix Microarray Suite Software were imported into the software dChip (50). All arrays were normalized against the array with median overall intensity. We used dChip to compute model-based expression indices (perfect match minus mismatch). To predict PTEN status on the basis of gene expression profiles, we used the RF predictors by Breiman (26). As a byproduct of its construction, an RF predictor yields an estimate of its error rate, a dissimilarity measure between the samples, and measures of variable importance. A measure of variable importance can be used to determine which genes are most important for predicting the tumor class. We chose the node purity-based importance measure (mean decrease in the Gini index). In the SI Appendix, we demonstrate that our findings are highly robust with respect to the choice of importance measure or choice of differential expression test. Specifically, we also considered the Kruskal–Wallis test, which is a nonparametric group comparison test and the Student t test of differential expression. As input of the classical multidimensional scaling plot (Fig. 1C) and average linkage hierarchical clustering analysis (Fig. 1D), we used the supervised RF dissimilarity measure that results from predicting PTEN status with the top 10 most important probe sets.
Serum from Mice Carrying Human Prostate and Breast Cancer Xenografts.
The animals were bled either retro-orbitally or by cardiac puncture. Blood samples were centrifuged at 2,000 × g for 5 min, and serum was collected and stored at −80 C.
Western Blot Analysis.
Lysates from the PTEN wild-type and deleted isogenic MEFs. MEF cells were lysed in RIPA buffer [0.15 mM NaCl/0.05 mM Tris·HCl (pH 7.2)/1% Triton X-100/1% sodium deoxycholate/0.1% SDS] plus protease/phosphatase inhibitors (Calbiochem, San Diego, CA) for 15 min on ice and sonicated at medium speed for five cycles of 30 seconds. Then they were centrifuged at full speed for 15 min at 4°C, and the supernatants were collected. Equal amounts of proteins in the concentrated samples were resolved by SDS/10% PAGE, electroblotted onto a nitrocellulose membrane (Invitrogen, Carlsbad, CA), blocked in 5% skim milk in 1× TBS, and probed with primary antibodies (PTEN, IGFBP2, β-actin, Akt, P-Akt, and HA-tag). Proteins were detected with an enhanced chemiluminescence kit (ECL; Amersham Pharmacia Biotech, Piscataway, NJ).
Tissue Culture.
The glioblastoma cell lines 9L (PTEN wild type) and U251 (PTEN mutant) cells were grown in DMEM (Cellgro, Herndon, VA) plus 10% FBS (Omega) medium. The prostate cancer cell lines LAPC4 (PTEN WT) and LNCaP (PTEN mutant) were grown in Iscove's modified Dulbecco's medium (Cellgro) plus 10% FBS. The breast cancer cell lines MDA-MB-468 and SKBR-3 cells were received from American Type Culture Collection (ATCC) (Manassas, VA). MDA-MB-468 cells were grown in DMEM: F12 media supplemented with 10% FBS. SKBR-3 cells are grown in DMEM supplemented with 10% FBS. PTEN+/+ and PTEN−/− MEFs were kindly provided by Hong Wu (University of California, Los Angeles) and were derived as described in ref. 27. Cells were transfected with the pcDNA 3.1 Zeo(+) plasmid encoding a constitutively active version of Akt (Invitrogen). IGFBP-2+/+ or IGFBP-2−/− MEFs were kindly provided by John Pintar (University of Medicine and Dentistry of New Jersey, Newark, NJ). Cells (5 × 104 cells per well) were plated in media containing 10% charcoal-stripped serum. Colonies were visualized by crystal violet staining and counted after 2 weeks of culture. The PI3-K inhibitor LY294002 was solubilized in DMSO and added in medium at the indicated concentrations.
Cell Cycle Assay.
Cells were trypsinized, fixed with 70% ethanol, and stained with propidium iodide by using a flow cytometry reagent set (Roche Applied Science, Indianapolis, IN) as described in ref. 51. Samples were analyzed for DNA content by using a FACScan flow cytometer and CellQuest software (Becton Dickinson, San Jose, CA).
Supplementary Material
Acknowledgments
We thank Hong Wu and John Pintar for providing the PTEN and IGFBP-2 MEFs, respectively; Tim Cloughesy and Paul Mischel for access to the University of California, Los Angeles, glioma tissues; and Chris Tran (Memorial Sloan–Kettering Cancer Center) and Igor Vivanco (University of California, Los Angeles) for their valuable contributions. J.K.V.R. is a Medical Foundation Fellow at the University of Sydney. C.L.S. received support as an Investigator of the Howard Hughes Medical Institute and as a Doris Duke Distinguished Clinical Scientist, as well as from a grant from the Prostate Cancer Foundation.
Abbreviations
- IGF1R
IGF1 receptor
- IGFBP2
insulin growth factor-binding protein 2
- MEF
mouse embryonic fibroblast
- PI3K
phosphatidylinositol 3-kinase
- RF
random forest.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609139104/DC1.
References
- 1.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 2.Ramaswamy S, Golub TR. J Clin Oncol. 2002;20:1932–1941. doi: 10.1200/JCO.2002.20.7.1932. [DOI] [PubMed] [Google Scholar]
- 3.Shai R, Shi T, Kremen TJ, Horvath S, Liau LM, Cloughesy TF, Mischel PS, Nelson SF. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 4.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 5.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, et al. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy S, Ross KN, Lander ES, Golub TR. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 7.Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, D'Amico M, Pestell RG, West M, Nevins JR. Nat Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- 8.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 9.Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, Zahnow CA, Patterson N, Golub TR, Ewen ME. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Coetzee GA. J Cell Biochem. 2004;93:233–241. doi: 10.1002/jcb.20228. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 12.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 13.Li DM, Sun H. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulis ML, Parsons R. Trends Cell Biol. 2003;13:478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 15.Paez J, Sellers WR. Cancer Treat Res. 2003;115:145–167. [PubMed] [Google Scholar]
- 16.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 17.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stambolic V, Tsao MS, Macpherson D, Suzuki A, Chapman WB, Mak TW. Cancer Res. 2000;60:3605–3611. [PubMed] [Google Scholar]
- 19.Maehama T, Dixon JE. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 20.Vivanco I, Sawyers CL. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 21.Baron-Hay S, Boyle F, Ferrier A, Scott C. Clin Cancer Res. 2004;10:1796–1806. doi: 10.1158/1078-0432.ccr-0672-2. [DOI] [PubMed] [Google Scholar]
- 22.Cohen P, Peehl DM, Stamey TA, Wilson KF, Clemmons DR, Rosenfeld RG. J Clin Endocrinol Metab. 1993;76:1031–1035. doi: 10.1210/jcem.76.4.7682560. [DOI] [PubMed] [Google Scholar]
- 23.Inman BA, Harel F, Audet JF, Meyer F, Douville P, Fradet Y, Lacombe L. Eur Urol. 2005;47:695–702. doi: 10.1016/j.eururo.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 24.Vorwerk P, Mohnike K, Wex H, Rohl FW, Zimmermann M, Blum WF, Mittler U. J Clin Endocrinol Metab. 2005;90:3022–3027. doi: 10.1210/jc.2004-0461. [DOI] [PubMed] [Google Scholar]
- 25.Whang YE, Wu X, Suzuki H, Reiter RE, Tran C, Vessella RL, Said JW, Isaacs WB, Sawyers CL. Proc Natl Acad Sci USA. 1998;95:5246–5250. doi: 10.1073/pnas.95.9.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breiman L. Neural Comput. 1999;11:1493–1517. doi: 10.1162/089976699300016106. [DOI] [PubMed] [Google Scholar]
- 27.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore MG, Wetterau LA, Francis MJ, Peehl DM, Cohen P. Int J Cancer. 2003;105:14–19. doi: 10.1002/ijc.11015. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Wang H, Shen W, Huang H, Hu L, Ramdas L, Zhou YH, Liao WS, Fuller GN, Zhang W. Cancer Res. 2003;63:4315–4321. [PubMed] [Google Scholar]
- 30.Rickman DS, Bobek MP, Misek DE, Kuick R, Blaivas M, Kurnit DM, Taylor J, Hanash SM. Cancer Res. 2001;61:6885–6891. [PubMed] [Google Scholar]
- 31.Canto EI, Shariat SF, Slawin KM. Urol Clin North Am. 2003;30:263–277. doi: 10.1016/s0094-0143(02)00183-0. [DOI] [PubMed] [Google Scholar]
- 32.Lai EC. Curr Biol. 2002;12:R74–78. doi: 10.1016/s0960-9822(01)00679-0. [DOI] [PubMed] [Google Scholar]
- 33.Maillard I, Pear WS. Cancer Cell. 2003;3:203–205. doi: 10.1016/s1535-6108(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 34.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, et al. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 35.Boulianne GL, de la Concha A, Campos-Ortega JA, Jan LY, Jan YN. EMBO J. 1991;10:2975–2983. doi: 10.1002/j.1460-2075.1991.tb07848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura H, Yoshida M, Tsuiki H, Ito K, Ueno M, Nakao M, Oka K, Tada M, Kochi M, Kuratsu J, et al. Oncogene. 1998;16:1009–1019. doi: 10.1038/sj.onc.1201618. [DOI] [PubMed] [Google Scholar]
- 38.Theodosiou A, Smith A, Gillieron C, Arkinstall S, Ashworth A. Oncogene. 1999;18:6981–6988. doi: 10.1038/sj.onc.1203185. [DOI] [PubMed] [Google Scholar]
- 39.Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 40.Popov SG, Krishna UM, Falck JR, Wilkie TM. J Biol Chem. 2000;275:18962–18968. doi: 10.1074/jbc.M001128200. [DOI] [PubMed] [Google Scholar]
- 41.Zumkeller W. Mol Pathol. 2001;54:285–288. doi: 10.1136/mp.54.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuller GN, Rhee CH, Hess KR, Caskey LS, Wang R, Bruner JM, Yung WK, Zhang W. Cancer Res. 1999;59:4228–4232. [PubMed] [Google Scholar]
- 43.Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi OP, Kononen J. Cancer Res. 2000;60:6617–6622. [PubMed] [Google Scholar]
- 44.Dahia PL. Endocr Relat Cancer. 2000;7:115–129. doi: 10.1677/erc.0.0070115. [DOI] [PubMed] [Google Scholar]
- 45.Levitt RJ, Georgescu MM, Pollak M. Biochem Biophys Res Commun. 2005;336:1056–1061. doi: 10.1016/j.bbrc.2005.08.229. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Chung D, Yang YC, Neely L, Tsurumoto S, Fan J, Zhang L, Biamonte M, Brekken J, Lundgren K, et al. Mol Cancer Ther. 2006;5:1256–1264. doi: 10.1158/1535-7163.MCT-05-0537. [DOI] [PubMed] [Google Scholar]
- 47.Bhat RV, Engber TM, Zhu Y, Miller MS, Contreras PC. J Pharmacol Exp Ther. 1997;281:522–530. [PubMed] [Google Scholar]
- 48.Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, Lamb DJ, Marcelli M, Belldegrun A, Witte ON, et al. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 49.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 50.Li C, Hung Wong W. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanazawa M, Satomi Y, Mizunati Y, Ukimura O, Kawauchi A, Sakai T, Baba M, Okuyama T, Nishino H, Miki T. Eur Urol. 2003;43:580–586. doi: 10.1016/s0302-2838(03)00090-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.