Abstract
Most studies of nucleotide binding to catalytic sites of Escherichia coli βY331W-F1-ATPase by the quenching of the βY331W fluorescence have been conducted in the presence of ≈20 mM sulfate. We find that, in the absence of sulfate, the nucleotide concentration dependence of fluorescence quenching induced by ADP, ATP, and MgADP is biphasic, revealing two classes of binding sites, each contributing about equally to the overall extent of quenching. For the high-affinity catalytic site, the Kd values for MgADP, ADP, and ATP equal 10, 43, and 185 nM, respectively. For the second class of sites, the Kd values for these ligands are ≈1,000× larger at 8.1, 37, and 200 μM, respectively. The presence of sulfate or phosphate during assay results in a marked increase in the apparent Kd values for the high-affinity catalytic site. The results show, contrary to earlier reports, that Mg2+ is not required for expression of different affinities for a nucleotide by the three catalytic sites. In addition, they demonstrate that the fluorescence of the introduced tryptophans is nearly completely quenched when only two sites bind nucleotide. Binding of ADP to the third site with a Kd near mM gives little fluorescence change. Many previous results of fluorescence quenching of introduced tryptophans appear to require reinterpretation. Our findings support a bi-site catalytic mechanism for F1-ATPase.
Keywords: ATP synthase, bi-site catalysis, phosphate, sulfate, tri-site catalysis
During oxidative and photophosphorylation, FoF1-ATP synthase is responsible for reversible synthesis of ATP coupled to transmembrane movement of H+ (or, in some bacteria, Na+). The enzyme can be separated into two multisubunit components, the membranous factor Fo, which in bacteria consists of a ring of c subunits and an a and b subunit in a stoichiometry of ab2cn, and the soluble factor F1 with a subunit composition of α3β3γδε. Energy transformation by FoF1 is best described by the binding change mechanism (1) and involves a rotation of a complex of subunits (γεcn, rotor) within the rest of the enzyme (stator) (2, 3). F1 is the catalytic component of ATP synthase and, when isolated, is capable only of net ATP hydrolysis. In the crystal structure of F1 from beef heart mitochondria (MF1), six nucleotide-binding sites are located at the interfaces between α and β subunits that are arranged alternately around an asymmetrical α-helical-coiled coil formed by γ-subunit (4). Three of the nucleotide-binding sites formed mostly by side chains of β subunits are catalytic, and another three primarily on α subunits are noncatalytic (5).
Three different conformations of β subunits in the crystal structure of MF1 are associated with asymmetric interactions with the γ subunit and are thought to represent distinct states that each of the catalytic sites sequentially assumes during catalytic cycle according to the binding change mechanism (4). That the γ subunit plays a crucial role in determining properties of the catalytic sites is supported by the crystal structure of α3β3-subcomplex of F1 from the thermophilic Bacillus PS3 (TF1). In this structure, all of the β subunits are in very similar conformations, and the three catalytic sites are in a state similar to one of the three states (open) found in the crystal structure of MF1 (6). In addition, the strong positive catalytic cooperativity in catalysis by F1 (7, 8) that is mediated by a rotation of γ subunit (9, 10) is lacking in the α3β3-subcomplex. Interactions between α and β subunits are also considered as contributing to the modulation of the state of catalytic sites in F1 (11, 12).
Whether asymmetric interactions of the three β subunits with γ, together with α/β interactions, are sufficient to induce asymmetry in the properties of the catalytic sites is not clear. According to Senior and colleagues (13–15), intersubunit interactions in F1 alone cannot induce nucleotide-binding heterogeneity at catalytic sites. Based on observations obtained with βY331W-mutant F1 from Escherichia coli (EcF1), it has been argued that in the absence of Mg2+, all three catalytic sites bind nucleotides with identical and low affinity and are in a similar state. Mg2+ is considered to play a crucial role in inducing asymmetry between catalytic sites, with high-affinity nucleotide binding at a single catalytic site. This role, according to (15), extends beyond Mg2+ being simply a required cofactor. However, this point of view is difficult to reconcile with the different reactivity toward chemical modifiers exhibited by each of the EcF1 catalytic sites in the absence of Mg2+ (16).
The catalytic site lacking bound nucleotide in the crystal structure of MF1 (17) and the catalytic sites in the crystal structure of the α3β3-subcomplex of TF1 (6) contain a bound sulfate ion. The presence of this sulfate in crystal structures suggests that the inclusion of sulfate in earlier nucleotide binding studies with βY331W-EcF1 (13–15) may have affected the results. In the present paper, we investigated nucleotide binding to βY331W-EcF1 in the presence and absence of Mg2+ and sulfate using the βY331W fluorescence as a signal. We used βY331W-EcF1 depleted of ε subunit (βY331W-εdEcF1) (18) to avoid heterogeneity in the enzyme forms during assay due to partial dissociation of ε.
The results show that, in the absence of sulfate, the concentration dependence of the fluorescence quenching induced in βY331W-εdEcF1 by ADP, ATP, and MgADP binding to the catalytic sites is clearly biphasic. One site with very high affinity and a second site with a much lower affinity contribute about equally to the overall extent of quenching. Sulfate reduces the affinity for nucleotide binding at the high-affinity catalytic site, leading to an increase in the apparent Kd. The results show that Mg2+ is not required for the asymmetry of nucleotide binding to the catalytic sites of the enzyme. They also show that most of the fluorescence of introduced tryptophans is quenched when two sites become occupied. Thus, the relationship between the extent of fluorescence quenching and nucleotide binding to all three sites is markedly nonlinear. These findings support a bi-site catalytic mechanism for F1-ATPases (19–23).
Results
Binding of ADP, MgADP, and ATP Without Sulfate Addition.
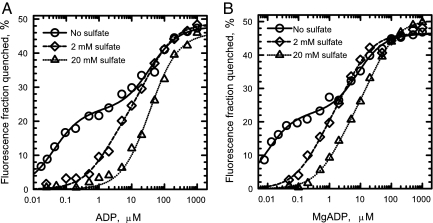
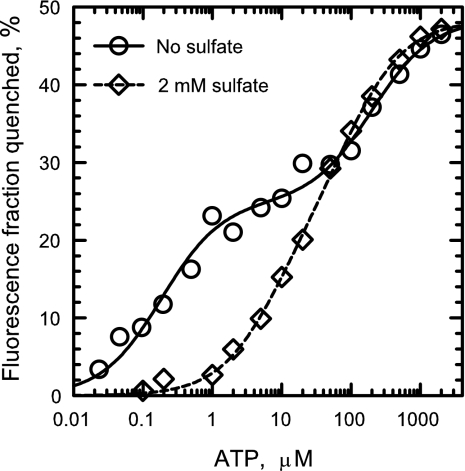
At saturating nucleotide concentrations, ADP (Fig. 1A), MgADP (Fig. 1B), and ATP (Fig. 2) binding to the catalytic sites of βY331W-εdEcF1 decreases the total fluorescence by ≈50%. This is similar to the results previously obtained both with ε-containing and ε-free forms of βY331W-EcF1 (18, 24, 25). However, in contrast to the data reported in ref. 25, nucleotide concentration dependence of the fluorescence quenching we obtained in the absence of Mg2+ was clearly biphasic (Figs. 1A and 2, circles). A model with two classes of independent binding sites (Eq. 2) satisfactorily fits the data (Figs. 1 and 2, solid lines), and the extent of quenching obtained upon filling the first and second sites (Q1 and Q2 from Eq. 2, respectively) presented in Table 1 indicates that nucleotide binding to each of the two classes of catalytic sites contributes about equally to the total fluorescence quenching. The Kd values for the higher affinity class of the catalytic sites (Kd,1) rise in the order MgADP < ADP < ATP (Table 1). The results obtained both with ADP (Fig. 1A, circles) and ATP (Fig. 2, circles) show that the catalytic sites of βY331W-εdEcF1 do exhibit asymmetry in nucleotide binding in the absence of Mg2+. Different affinities of the catalytic sites for a nucleotide in the absence of Mg2+ have also been observed with the homologous βY341W-mutant α3β3γ-subcomplex of TF1 (26). We did not attempt to measure Kd values for MgATP as has been done previously (18, 27) because the rapid formation of catalytic site ADP makes it uncertain what nucleotide affinity is being measured.
Fig. 1.
Effect of sulfate on ADP and MgADP binding to the catalytic sites of βY331W-εdEcF1. Fluorescence quenching induced by ADP (A) and MgADP (B) was measured in MTE buffer (A) and MTE buffer containing additional 3.2 mM MgAc2 (B) as described under Experimental Procedures in the absence (circles) and presence of 2 mM (diamonds) and 20 mM (triangles) K2SO4. Solid and dashed lines in A and solid, dashed, and dotted lines in B represent the best fit of the data to Eq. 2, and dotted line in A represents the best fit of the data to Eq. 3, with the results of the fit shown in Table 1.
Fig. 2.
Effect of sulfate on ATP binding to the catalytic sites of βY331W-εdEcF1. ATP-induced fluorescence quenching was measured in MTE buffer as described under Experimental Procedures in the absence (circles) and presence of 2 mM K2SO4 (diamonds). Solid and dashed lines represent the best fit of the data to Eq. 2, with the results of the fit shown in Table 1.
Table 1.
Best-fit parameters of the nucleotide-induced fluorescence quenching in βY331W-εdEcF1
| Nucleotide | Additions |
Estimated |
||||
|---|---|---|---|---|---|---|
| Sulfate, mM | Phosphate, mM | Kd,1, μM | Q1, % | Kd,2, μM | Q2, % | |
| ADP* | − | − | 0.043 ± 0.007 | 23.4 ± 0.9 | 37 ± 8 | 24.4 ± 1.2 |
| ADP* | 2 | − | 1.8 ± 0.6 | 21.7 ± 4 | 36 ± 12 | 27 ± 4 |
| ADP† | 20 | − | 37 ± 3‡ | 46 ± 1§ | ||
| ADP* | − | 5 | 2.4 ± 0.6 | 27.1 ± 3.5 | 60 ± 20 | 25.5 ± 3.2 |
| MgADP* | − | − | 0.01 ± 0.002 | 22.7 ± 0.7 | 8.1 ± 1.3 | 23.7 ± 0.8 |
| MgADP* | 2 | − | 0.35 ± 0.2 | 21.8 ± 8 | 4.7 ± 2.6 | 24.7 ± 8 |
| MgADP* | 20 | − | 1.8 ± 0.3 | 22.8 ± 2.5 | 28 ± 5 | 27.2 ± 2.4 |
| ATP* | − | − | 0.185 ± 0.03 | 25 ± 1 | 200 ± 60 | 23.4 ± 1.8 |
| ATP* | 2 | − | 7.5 ± 2 | 22.5 ± 5 | 100 ± 30 | 25.8 ± 4.6 |
Effect of Sulfate and Phosphate on Binding Patterns.
As noted earlier, crystal structures suggest that sulfate is present at catalytic sites and could interfere with nucleotide binding. The results shown in Figs. 1 (diamonds and triangles) and 2 (diamonds) demonstrate that sulfate indeed can markedly increase the Kd,1 values for ADP, MgADP, and ATP for binding to the high-affinity catalytic site. In the absence of nucleotide, 2 and 20 mM sulfate increased the fluorescence of βY331W-εdEcF1 by ≈5% and 10%, respectively (for comparison, 20 mM sulfate increased fluorescence of the wild-type εdEcF1 by ≈2%). In the presence of 2 mM sulfate, the asymmetry of ADP, MgADP, and ATP binding to the catalytic sites of βY331W-εdEcF1 became less noticeable (diamonds in Figs. 1 A and B and 2, respectively) largely due to an increase in the apparent Kd,1 values (Table 1).
Triangles in Fig. 1A show the ADP-dependence of βY331W-εdEcF1 fluorescence quenching obtained when sulfate concentration was increased to 20 mM. In this case, the model with one class of binding sites (Eq. 3) satisfactorily fits the data with the Kd and Qt values of 36 μM and 46%, respectively (Table 1 and Fig. 1A, dotted line). When βY331W-EcF1 was titrated by Löbau et al. (25) with ADP in a medium containing 50 mM Tris/SO4 buffer at pH 8 (sulfate concentration of ≈17 mM) (figure 3B in ref. 25), a single Kd was sufficient to fit the data. A single apparent Kd of 63 and 85 μM for ADP binding to βY331W-εdEcF1 under the same conditions was reported in the absence and presence of ε, respectively (18).
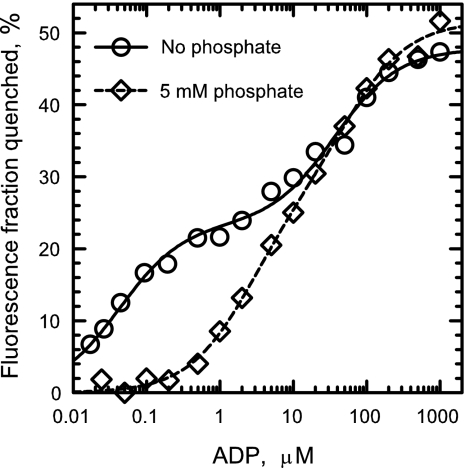
Fig. 3.
Effect of phosphate on ADP binding to the catalytic sites of βY331W-εdEcF1. ADP-induced fluorescence quenching was measured in MTE buffer as described under Experimental Procedures in the absence (circles) and presence of 5 mM phosphate (diamonds). The data obtained in the absence of phosphate (circles) and the corresponding solid line are from Fig. 1A. Dashed line represents the best fit of the data to Eq. 2 with the results of the fit shown in Table 1.
In addition to decreasing the affinity of nucleotide binding to the high-affinity site, sulfate may also reduce, although to a lesser extent, the affinity of MgADP for binding to the lower affinity class of catalytic sites on βY331W-εdEcF1 (Fig. 1B, triangles). In the presence of 20 mM sulfate, the apparent Kd,1 and Kd,2 values for MgADP of 1.8 and 28 μM, respectively, obtained in the present study (Table 1 and Fig. 1B, triangles and dotted line) are close to the values (0.7 and 25 μM, respectively) obtained earlier with βY331W-εdEcF1 under similar conditions (18).
Similar to sulfate, Pi also significantly inhibits quenching of the βY331W-εdEcF1 fluorescence induced by ADP at lower concentrations (Fig. 3, diamonds). The results of the best fit to Eq. 2 (Fig. 3, dashed line, and Table 1) show that Pi reduces the apparent ADP affinity for binding to the high-affinity catalytic site. As shown in Table 1, the assumption that quenching in the absence of sulfate or phosphate is governed by the filling of two classes of nucleotide-binding sites of equal abundance differing in affinity ≈1,000-fold is well supported by the results.
Discussion
Fluorescence Properties of Tryptophan in Proteins.
For discussion of the results, it is important to point out that the quantum yield for tryptophan fluorescence in proteins may vary from near zero to ≈0.35 (28). In EcF1, βY331W residues have been estimated to have an average quantum yield of 0.13 (29). This means that fluorescence of these residues is significantly quenched even in the absence of bound nucleotides. As noted later in this discussion, results presented here and other published results give evidence that the fluorescence of the inserted tryptophans differs considerably.
Asymmetry of Nucleotide Binding in the Absence of Mg2+.
The nucleotide concentration dependence of the βY331W-εdEcF1 fluorescence quenching in the absence of Mg2+ (Table 1 and Figs. 1A and 2, circles) shows that at least two of the three catalytic sites exhibit significantly different affinities to a nucleotide. These results show that Mg2+ is not required for expression of an asymmetry in the nucleotide-binding properties by the catalytic sites of EcF1. In the absence of sulfate, the high-affinity catalytic site in βY331W-εdEcF1 binds MgADP with a Kd of 10 nM (Table 1) and ADP with a Kd of ≈50 nM (Table 1). The results dispute the notion (13–15) that Mg2+ plays a special role in, and is obligatory for, the asymmetry of the catalytic sites on EcF1. It appears that the catalytic-site asymmetry in F1, including the capacity to express the high-affinity catalytic site, is determined primarily by nonequivalent interactions of the three β subunits with the single γ subunit.
The Marked Decrease in Nucleotide Affinity Caused by Sulfate.
Results presented in Figs. 1 and 2 show that 2 mM sulfate increases the values of Kd,1 for ADP, MgADP, and ATP binding to the higher affinity class of the catalytic sites in βY331W-εdEcF1 without appreciably affecting the values of Kd,2 and the corresponding Q1 and Q2 values (Table 1). Further increases in Kd,1 values for ADP and MgADP are observed with the addition of 20 mM sulfate (Fig. 1, triangles). With 20 mM sulfate, only one Kd value, which is practically undistinguished from the Kd,2 values obtained in the absence or presence of 2 mM sulfate, gives a good fit to the data obtained with ADP (Table 1 and Fig. 1A, dotted line). The ability of sulfate to decrease the affinity of βY331W-εdEcF1 catalytic sites for nucleotides is likely responsible for removal of endogenous nucleotides from the catalytic sites of the enzyme during column centrifugation (30) in the presence of sulfate (24).
Bandyopadhyay et al. (26) have suggested that endogenous nucleotides bound at the noncatalytic sites of βY331W-EcF1 are responsible for the apparent lack of asymmetry in the nucleotide binding in the absence of Mg2+ reported earlier (24). Our results make it more likely that the main reason that the catalytic-site asymmetry had not been detected in the absence of Mg2+ was the presence of ≈17 mM sulfate that competed with nucleotide binding to the high-affinity site.
In the crystal structure of the α3β3-subcomplex of TF1 (6), sulfate at the catalytic sites occupies a position close to that of the β-phosphate of nucleotide bound at the catalytic site in the crystal structure of MF1 (4). An electron density that is consistent with a bound sulfate ion has also been found in the open catalytic site in the crystal structure of MF1 (17). This density is located in a region of the P-loop residues that are involved in interactions with the phosphoryl groups of bound nucleotide (ADP and AMPPNP) in occupied catalytic sites (17). Such sulfate binding would be expected to reduce the ADP and ATP binding at the catalytic sites.
The cause of the increase in fluorescence when sulfate binds in the absence of nucleotide is uncertain. The inserted tryptophans at sites 1 and 2 already show considerable quenching, and this may be partially relieved by the sulfate.
Effect of Phosphate on Nucleotide Affinity.
Similarly to sulfate, Pi reduces ADP binding to the high-affinity catalytic site in βY331W-εdEcF1 (Fig. 3 and Table 1). Pi binding with Kd values in the range of 100 μM for at least two sites has been recently detected in EcF1 in the presence of Mg2+ using pressure dialysis (31). No competing effects of Pi on nucleotide binding to catalytic sites monitored by the fluorescence quenching were reported with βY331W-EcF1 in the presence of Mg2+ (27, 32) and with βY331W-EcFoF1 in the absence of Mg2+ (33). This is likely due to the presence of ≈20 mM sulfate in these experiments, which would have already reduced nucleotide affinity significantly.
Site Filling and Fluorescence Quenching.
Without sulfate present, the two-component nature of the ADP and MgADP concentration dependence of quenching of nearly all of the extra fluorescence of βY331W-εdEcF1 is clearly evident (Figs. 1 and 2 and Table 1). Both components have approximately equal amplitudes. The binding at one high-affinity site and at a second site with Kd values ≈30–70 μM have been reported by others to result in quenching of nearly all of the extra fluorescence. This includes titration of βY331W-EcF1 fluorescence with MgADP (25, 27, 29, 34) and MgAMPPNP (27, 32) and titration of the homologous βY341W-mutant α3β3γ-subcomplex of TF1 with MgADP and MgATP (26, 35, 36). Each component could logically represent binding at only one catalytic site. This would mean that nearly all observed quenching occurs when only two sites are filled. An alternate interpretation has been that one of the components reflects binding at two catalytic sites with close or identical affinities for a nucleotide. Such an explanation has been suggested for interpreting the results obtained during titration of βY331W-EcF1 fluorescence with MgADP (25, 27, 29, 34) and MgAMPPNP (27, 32). However, the occurrence of nearly identical affinities seems unlikely in view of the well documented differences in the structure and chemical properties of the three catalytic sites (see refs. 20 and 37 for review). Our and other results are consistent with a requirement of only filling of two sites for the near total fluorescence quenching. This would mean that the tryptophan present at a third site must contribute little to the extra fluorescence from the inserted tryptophans, and if it binds nucleotide at a higher concentration, this is not reflected in a fluorescence change. That this somewhat unexpected behavior is what actually occurs is considered in the following section.
Nucleotide Binding at a Third Site Occurs with Little or No Fluorescence Quenching.
The possibility that all three catalytic sites fill with MgADP with Kd values <100 μM is rendered unlikely by recent results obtained by Ahmad and Senior (38–41). The authors investigated the efficiency of MgADP in protecting one of the catalytic sites from modification by NBD-Cl (42–46) in wild-type EcF1 and in a range of double-mutant EcF1 containing the βY331W substitution. The half-time of the wild-type EcF1 inactivation by 0.1 mM NBD-Cl was increased from ≈7 min in the absence of the nucleotide (figure 8A in ref. 39) to 60 min in the presence of ≈4 mM MgADP (figure 7A in ref. 39). The results lead to an apparent Kd value for MgADP of ≈0.5 mM. This value is significantly higher than the apparent Kd value of 35–40 μM for MgADP that have been obtained for the lowest affinity catalytic site in βY331W-EcF1 under identical conditions using nucleotide-induced fluorescence quenching (39, 40).
In addition, results obtained with EcF1 containing double mutations βY331W/βR246Q, βY331W/βR246K, and βY331W/βR246A (39); βY331W/βN243A (40), βY331W/αR376K, and βY331W/αR376Q (41, 47); and βY331W/βR182K (41, 48) allow estimation of Kd values for MgADP from the nucleotide-conferred protection against inactivation by NBD-Cl. The values are 7- to 180-fold higher than those reported for the catalytic site, with the lowest affinity to the nucleotide using the fluorescence quenching method. These results mean that MgADP binding to the third catalytic site in βY331W-EcF1 takes place after the fluorescence of the three βY331W residues has already been quenched almost completely or completely due to nucleotide binding to the first and second catalytic sites. Such findings warrant the conclusion that nucleotide binding at a third site has little effect on fluorescence quenching.
That nucleotide binding at a third catalytic site with a Kd in the mM range is not associated with a change in fluorescence is also shown by the lack of increased quenching as >1 mM nucleotide concentrations are approached (Figs. 1 and 2). The lack of fluorescence quenching associated with nucleotide binding at a third site, and the practically complete quenching of fluorescence of the three βY331W residues at saturating nucleotide concentrations (see also refs. 18 and 27), lead to the conclusion that the extent of quenching reflects the occupancy of two, and not three, catalytic sites. As noted earlier, we conclude that these results support the bi-site mechanism suggested for the F1-ATPases.
Such a conclusion harmonizes with results obtained when the double αW463F/βY341W-mutant α3β3γ-subcomplex of TF1 was titrated with MgATP (49). They strongly suggested unequal contributions from each of the three catalytic sites to either the initial fluorescence or the nucleotide-induced fluorescence quenching, or to both.
Other Evidence Supporting Bi-Site Catalysis.
It warrants mention that the bi-site mechanism is also supported by measurements of the catalytic site occupancy during steady-state MgATP hydrolysis by MF1 (23) and EcF1 (Y.M.M., V.V.B., and Richard L. Cross, unpublished data). In these studies, the amount of bound nucleotide was measured using a centrifugal filtration method. They show that, as ATP concentration is increased, rapid catalytic turnover occurs as two catalytic sites become filled with nucleotide.
In contrast, fluorescence quenching data obtained with βY331W-EcF1 during MgATP hydrolysis, with a single Km of 20–40 μM (18, 27, 50), have been interpreted as showing that rapid catalytic turnover occurs only when three catalytic sites have been filled with nucleotide (13, 15, 27). This and most earlier interpretations based on the quenching of the fluorescence of introduced tryptophans appear to need reconsideration.
Experimental Procedures
Enzyme Preparation.
To generate βY331W mutation (27), a mutant construct p3UβY331W was obtained using mutagenic oligonucleotide 5′-TGGGTATCTGGCCGGCCGTT, with bases in italic generating the mutation and p3U+ plasmid (51) as a template and expressed in JP17 strain (52).
βY331W-EcF1 was purified according to ref. 53, with the modifications described in ref. 9. βY331W-EcF1 depleted of ε subunit (βY331W-εdEcF1) was prepared as described (54). Before use, βY331W-εdEcF1 was treated to remove nucleotides bound at the catalytic sites as described (24) with the following modifications. βY331W-εdEcF1 was diluted to a concentration of 4.5 μM by a buffer composed of 20 mM Mops/Tris, pH 8.0, 150 mM sucrose, 0.2 mM EDTA (MTSE buffer) and containing additionally 20 mM K2SO4. After a 10-min incubation at room temperature (22°C), the enzyme was centrifuged through a column (30) equilibrated with MTSE buffer containing additionally 20 mM K2SO4. The column effluent was incubated for 20 min and subjected to a second centrifugation through a column equilibrated with MTSE buffer containing additionally 20 mM K2SO4. After 20-min incubation, sulfate was removed from the enzyme solution using two centrifugations through columns equilibrated with MTSE buffer and a 20-min incubation period between the centrifugations.
Fluorescence Measurements.
Fluorescence was measured using a FluoroLog FL3-21 spectrofluorometer (Jobin Yvon) at 22°C in 1 cm × 1 cm quartz cuvettes under constant stirring. The excitation wavelength was 295 nm (2-nm bandpass), and the emission wavelength was 360 nm (10-nm bandpass). The fluorescence assay (final volume 2 ml) was initiated by adding βY331W-εdEcF1 (final concentration 7–8 nM) into a medium containing 20 mM Mops/Tris (pH 8.0) and 0.2 mM EDTA (MTE buffer) and nucleotides and anions as specified in the figure legends. The fluorescence intensity was measured 10 min later. Where indicated, the assay medium contained an additional 3.2 mM MgAc2. The extent of the fluorescence quenching (Q) was calculated according to
where F0 and Fn are the fluorescence intensities in the absence and presence of a nucleotide after subtracting the background signals, and kif is the factor correcting for the nucleotide-induced inner filter effect. Values of kif were obtained using N-acetyl-l-tryptophanamide.
Eqs. 2 and 3 were used to analyze the data using SigmaPlot 8.0 (SPSS, Chicago, IL):
where c is the free nucleotide concentration, and Q1, Q2, and Qt are the extent of the fluorescence quenching produced by nucleotide binding to the sites characterized by Kd,1, Kd,2, and Kd, respectively. Eq. 2 represents a model with two classes of the independent binding sites, and Eq. 3 represents a model with one class of the binding sites. The value of c was obtained by subtracting the bound nucleotide concentration from the added nucleotide concentration. The bound nucleotide concentration was calculated from the extent of quenching assuming that quenching at saturating nucleotide concentration corresponded to the filling of three catalytic sites.
Acknowledgments
We thank Drs. P. D. Boyer and R. L. Cross for valuable encouragement and discussions. This work was supported in part by National Institutes of Health Grant GM23152 (to R. L. Cross).
Abbreviations
- EcF1
solubilized ATPase portion of FoF1-ATP synthase from Escherichia coli
- MF1
solubilized ATPase portion of FoF1-ATP synthase from beef heart mitochondria
- AMPPNP
5′-adenylyl-β,γ-imidodiphosphate
- NBD-Cl
7-chloro-4-nitrobenzo-2-oxa-1,3-diazole
- TF1
solubilized ATPase portion of FoF1-ATP synthase from thermophilic Bacillus PS3.
Footnotes
The authors declare no conflict of interest.
References
- 1.Boyer PD. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 2.Kinosita K, Jr, Yasuda R, Noji H, Adachi K. Philos Trans R Soc London B. 2000;355:473–489. doi: 10.1098/rstb.2000.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stock D, Gibbons C, Arechaga I, Leslie AGW, Walker JE. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 4.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 5.Cross RL, Nalin CM. J Biol Chem. 1982;257:2874–2881. [PubMed] [Google Scholar]
- 6.Shirakihara Y, Leslie AGW, Abrahams JP, Walker JE, Ueda T, Sekimoto Y, Kambara M, Saika K, Kagawa Y, Yoshida M. Structure (London) 1997;5:825–836. doi: 10.1016/s0969-2126(97)00236-0. [DOI] [PubMed] [Google Scholar]
- 7.Grubmeyer C, Cross RL, Penefsky HS. J Biol Chem. 1982;257:12092–12100. [PubMed] [Google Scholar]
- 8.Cross RL, Grubmeyer C, Penefsky HS. J Biol Chem. 1982;257:12101–12105. [PubMed] [Google Scholar]
- 9.Duncan TM, Bulygin VV, Zhou Y, Hutcheon ML, Cross RL. Proc Natl Acad Sci USA. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 11.Ren H, Allison WS. Biochim Biophys Acta. 2000;1458:221–233. doi: 10.1016/s0005-2728(00)00075-x. [DOI] [PubMed] [Google Scholar]
- 12.Futai M, Omote H, Sambongi Y, Wada Y. Biochim Biophys Acta. 2000;1458:276–288. doi: 10.1016/s0005-2728(00)00080-3. [DOI] [PubMed] [Google Scholar]
- 13.Weber J, Senior AE. Biochim Biophys Acta. 1997;1319:19–58. doi: 10.1016/s0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 14.Senior AE, Nadanaciva S, Weber J. J Exp Biol. 2000;203:35–40. doi: 10.1242/jeb.203.1.35. [DOI] [PubMed] [Google Scholar]
- 15.Senior AE, Nadanaciva S, Weber J. Biochim Biophys Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 16.Haughton MA, Capaldi RA. J Biol Chem. 1995;270:20568–20574. doi: 10.1074/jbc.270.35.20568. [DOI] [PubMed] [Google Scholar]
- 17.Menz RI, Leslie AGW, Walker JE. FEBS Lett. 2001;494:11–14. doi: 10.1016/s0014-5793(01)02302-x. [DOI] [PubMed] [Google Scholar]
- 18.Weber J, Dunn SD, Senior AE. J Biol Chem. 1999;274:19124–19128. doi: 10.1074/jbc.274.27.19124. [DOI] [PubMed] [Google Scholar]
- 19.Zhou JM, Boyer PD. J Biol Chem. 1993;268:1531–1538. [PubMed] [Google Scholar]
- 20.Boyer PD. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 21.Milgrom YM, Murataliev MB, Boyer PD. Biochem J. 1998;330:1037–1043. doi: 10.1042/bj3301037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer PD. FEBS Lett. 2002;512:29–32. doi: 10.1016/s0014-5793(02)02293-7. [DOI] [PubMed] [Google Scholar]
- 23.Milgrom YM, Cross RL. Proc Natl Acad Sci USA. 2005;102:13831–13836. doi: 10.1073/pnas.0507139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber J, Wilke-Mounts S, Senior AE. J Biol Chem. 1994;269:20462–20467. [PubMed] [Google Scholar]
- 25.Löbau S, Weber J, Wilke-Mounts S, Senior AE. J Biol Chem. 1997;272:3648–3656. doi: 10.1074/jbc.272.6.3648. [DOI] [PubMed] [Google Scholar]
- 26.Bandyopadhyay S, Valder CR, Huynh HG, Ren H, Allison WS. Biochemistry. 2002;41:14421–14429. doi: 10.1021/bi026243g. [DOI] [PubMed] [Google Scholar]
- 27.Weber J, Wilke-Mounts S, Lee RS-F, Grell E, Senior AE. J Biol Chem. 1993;268:20126–20133. [PubMed] [Google Scholar]
- 28.Chen Y, Barkley MD. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 29.Weber J, Senior AE. J Biol Chem. 1998;273:33210–33215. doi: 10.1074/jbc.273.50.33210. [DOI] [PubMed] [Google Scholar]
- 30.Penefsky HS. J Biol Chem. 1977;252:2891–2899. [PubMed] [Google Scholar]
- 31.Penefsky HS. FEBS Lett. 2005;579:2250–2252. doi: 10.1016/j.febslet.2005.02.072. [DOI] [PubMed] [Google Scholar]
- 32.Weber J, Senior AE. J Biol Chem. 1995;270:12653–12658. doi: 10.1074/jbc.270.21.12653. [DOI] [PubMed] [Google Scholar]
- 33.Löbau S, Weber J, Senior AE. Biochemistry. 1998;37:10846–10853. doi: 10.1021/bi9807153. [DOI] [PubMed] [Google Scholar]
- 34.Nadanaciva S, Weber J, Senior AE. J Biol Chem. 1999;274:7052–7058. doi: 10.1074/jbc.274.11.7052. [DOI] [PubMed] [Google Scholar]
- 35.Dou C, Fortes PAG, Allison WS. Biochemistry. 1998;37:16757–16764. doi: 10.1021/bi981717q. [DOI] [PubMed] [Google Scholar]
- 36.Ren H, Allison WS. J Biol Chem. 2000;275:10057–10063. doi: 10.1074/jbc.275.14.10057. [DOI] [PubMed] [Google Scholar]
- 37.Walker JE. Angew Chem Int Ed. 1998;37:2309–2319. [Google Scholar]
- 38.Ahmad Z, Senior AE. J Bioenerg Biomembr. 2005;37:437–440. doi: 10.1007/s10863-005-9486-8. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad Z, Senior AE. J Biol Chem. 2004;279:31505–31513. doi: 10.1074/jbc.M404621200. [DOI] [PubMed] [Google Scholar]
- 40.Ahmad Z, Senior AE. J Biol Chem. 2004;279:46057–46064. doi: 10.1074/jbc.M407608200. [DOI] [PubMed] [Google Scholar]
- 41.Ahmad Z, Senior AE. FEBS Lett. 2005;579:523–528. doi: 10.1016/j.febslet.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 42.Ferguson SJ, Lloyd WJ, Lyons MH, Radda GK. Eur J Biochem. 1975;54:117–126. doi: 10.1111/j.1432-1033.1975.tb04120.x. [DOI] [PubMed] [Google Scholar]
- 43.Lunardi J, Satre M, Bof M, Vignais PV. Biochemistry. 1979;18:5310–5316. doi: 10.1021/bi00591a008. [DOI] [PubMed] [Google Scholar]
- 44.Andrews WW, Hill FC, Allison WS. J Biol Chem. 1984;259:8219–8225. [PubMed] [Google Scholar]
- 45.Sutton R, Ferguson SJ. Eur J Biochem. 1985;148:551–554. doi: 10.1111/j.1432-1033.1985.tb08875.x. [DOI] [PubMed] [Google Scholar]
- 46.Orriss GL, Leslie AGW, Braig K, Walker JE. Structure (London) 1998;6:831–837. doi: 10.1016/s0969-2126(98)00085-9. [DOI] [PubMed] [Google Scholar]
- 47.Nadanaciva A, Weber J, Wilke-Mounts S, Senior AE. Biochemistry. 1999;38:15493–15499. doi: 10.1021/bi9917683. [DOI] [PubMed] [Google Scholar]
- 48.Nadanaciva A, Weber J, Senior AE. Biochemistry. 1999;38:7670–7677. doi: 10.1021/bi990663x. [DOI] [PubMed] [Google Scholar]
- 49.Ono S, Hara KY, Hirao J, Matsui T, Noji H, Yoshida M, Muneyuki E. Biochim Biophys Acta. 2003;1607:35–44. doi: 10.1016/j.bbabio.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Weber J, Senior AE. J Biol Chem. 2001;276:35422–35428. doi: 10.1074/jbc.M104946200. [DOI] [PubMed] [Google Scholar]
- 51.Duncan TM, Zhou Y, Bulygin VV, Hutcheon ML, Cross RL. Biochem Soc Trans. 1995;23:736–741. doi: 10.1042/bst0230736. [DOI] [PubMed] [Google Scholar]
- 52.Lee RS-F, Pagan J, Wilke-Mounts S, Senior AE. Biochemistry. 1991;30:6842–6847. doi: 10.1021/bi00242a006. [DOI] [PubMed] [Google Scholar]
- 53.Senior AE, Downie JA, Cox GB, Gibson F, Langman L, Fayle DRH. Biochem J. 1979;180:103–109. doi: 10.1042/bj1800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunn SD. Anal Biochem. 1986;159:35–42. doi: 10.1016/0003-2697(86)90304-0. [DOI] [PubMed] [Google Scholar]