Abstract
The genetic factors contributing to the complex disorder of myocardial calcification are largely unknown. Using a mouse model, we fine-mapped the major locus (Dyscalc1) contributing to the dystrophic cardiac calcification (DCC) to an 840-kb interval containing 38 genes. We then identified the causal gene by using an approach integrating genetic segregation and expression array analyses to identify, on a global scale, cis-acting DNA variations that perturb gene expression. By studying two intercrosses, in which the DCC trait segregates, a single candidate gene (encoding the ATP-binding cassette transporter ABCC6) was identified. Transgenic complementation confirmed Abcc6 as the underlying causal gene for Dyscalc1. We demonstrate that in the cross, the expression of Abcc6 is highly correlated with the local mineralization regulatory system and the BMP2-Wnt signaling pathway known to be involved in the systemic regulation of calcification, suggesting potential pathways for the action of Abcc6 in DCC. Our results demonstrate the power of the integrative genomics in discovering causal genes and pathways underlying complex traits.
Keywords: expression quantitative trait locus, transgenic, positional cloning, osteopontin
Characterized by hydroxyapatite deposition in necrotic myocytes, myocardial calcification is common in specific forms of cardiomyopathy and in myocardial infarction. It has been estimated that ≈8% of patients with severe myocardial infarction develop myocardial calcification within 6 years, suggesting a genetic predisposition for postinjury healing and remodeling processes (1). Historically, dystrophic cardiac calcification (DCC) has been considered a spontaneous form of cardiomyopathy in mice, associated with a variety of predisposing factors, but with normal blood levels of calcium and phosphate. Experimentally, it can be reproducibly initiated using a transdiaphragmal freeze–thaw injury or a high-phosphorous (HP) diet (2, 3). Several inbred mouse strains, including C3H/HeJ (C3H) and DBA/2J (DBA), are highly susceptible, whereas many other inbred mouse strains, including C57BL/6J (B6), C57BL/10J (B10), A/J, MRL/MpJ, and BALB/cJ are resistant (refs. 4 and 5; X.W., T.A.D., and A.J.L., unpublished data). In DCC susceptible strains, calcification has also been observed in skeletal muscle, including in the tongue and diaphragm, and kidney, suggesting a systemic defect (2, 5).
Using quantitative trait locus (QTL) analysis of an F2 intercross between B6 and C3H mice (BxH), we previously mapped four DCC loci (6, 7). The locus on chromosome 7 (Dyscalc1), which exhibits recessive inheritance, explains 31% of the total genetic variance and is the major contributor. Dyscalc1 was confirmed by separate intercrosses of B6 and DBA mice (BxD) (5, 8). The C3H strain was originally derived from an outbreeding experiment of the DBA strain, leading us to hypothesize that the susceptible strains C3H and DBA share a common disease-causing allele (8). To fine map the Dyscalc1 locus, we screened a panel of recombinant congenic (RC) strains generated from a B10 donor strain on a C3H recipient genetic background (9). Of the 16 RC strains screened, three (HcB12, HcB24, and HcB28) were found to display the DCC-resistant phenotype. Using markers from the Dyscalc1 linkage peak region, we detected B10 homozygote alleles in both the HcB12 and HcB24 strains, thus reducing the Dyscalc1 locus to a 7-cM interval on chromosome 7 (8).
To identify the causal gene underlying Dyscalc1 and to begin investigating the disease mechanism of DCC, we took advantage of the genome-wide expression array data generated in two intercrosses. We used an integrative genomics approach to identify Abcc6 as the major gene contributing to DCC in mice. We further demonstrated that Abcc6 exhibits coexpression in response to genetic perturbations with several other genes in pathways known to be involved in systemic calcification.
Results
Fine Mapping of Dyscalc1.
We generated a backcross population between strains HcB24 and C3H (HcB24xC3H) and screened N2 mice for recombinant progeny in the 7-cM interval. With >93% of the genome derived from the C3H genetic background, and all three other DCC controlling loci fixed for their C3H alleles, the Dyscalc1 locus exhibited full penetrance. A total of 649 backcross mice were generated, yielding 49 recombinants. Genotypic and phenotypic characterization of a total of 23 recombinant progeny within the congenic interval restricted the Dyscalc1 locus to an 840-kb region containing 38 genes [supporting information (SI) Fig. 5].
Identification of Abcc6 as a Candidate Gene.
We hypothesized that the gene for DCC at the chromosome 7 locus was likely to be differentially expressed between the susceptible and resistant strains because of a cis-acting variation. To test this, we examined existing expression array data generated for progeny of intercrosses of BxH (10) and BxD (11) mice (SI Methods). The BxD data set contains liver array data from 111 F2 mice, whereas the BxH data set contains array data of 334 F2 mice from liver, skeletal muscle, adipose tissue, and brain. Although the primary calcification phenotype occurs in myocardium and heart is our main tissue of interest, we used array data from these other tissues to perform gene expression profiling analyses. Our data suggest that for a gene exhibiting significant cis-regulation in one tissue, there is a 63–88% probability that it also exhibits cis-regulation in another tissue if it is actively expressed in that tissue (SI Table 1) (12). We treated the liver expression data of each gene in the databases as a quantitative trait to study linkage in a segregating F2 population. In each cross, thousands of QTLs for transcript levels (expression QTLs or eQTLs) were identified, and those eQTLs mapping over their corresponding structural genes were considered to be cis-acting (i.e., because of variations of the gene itself). This cis-eQTL approach to identify cis-acting genes has been previously validated using a classic cis-trans test (13). Because both the BxH and BxD intercrosses demonstrated a DCC QTL at the Dyscalc1 locus, we reasoned that the causal gene would exhibit a cis-eQTL in both crosses. Of the 34 genes represented on the microarray, eight had significant cis-eQTL [logarithm of odds (lod) >4.3] in the BxH cross and two in the BxD cross using the liver data set (SI Table 2). Only the ATP-binding cassette transporter gene, Abcc6, exhibited significant cis-eQTL lod scores in both crosses. Similarly, we studied microarray data from the BxH muscle, adipose, and brain data sets and found that only Abcc6 expression, specifically liver expression levels of Abcc6, correlated significantly with both myocardial calcification (P < 0.0001) and aortic medial calcification (P = 0.0007) in the BxH cross. This finding is consistent with our hypothesis and also suggests a systemic impact of Abcc6 expression in liver. Overall, these results identified Abcc6 as a strong candidate gene for Dyscalc1.
Expression and Sequence Variations of Abcc6 in DBA, C3H, and B6.
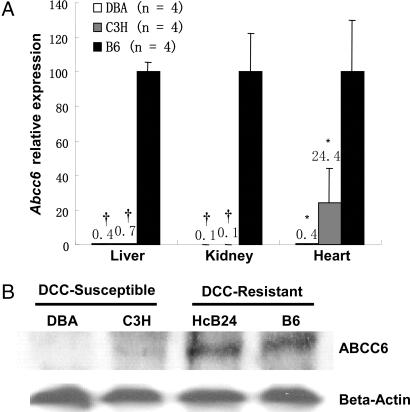
Quantitative RT-PCR (QPCR) analysis confirmed dramatically reduced expression of Abcc6 in strains C3H and DBA as compared with B6 in liver, kidney, and heart (Fig. 1A and SI Table 3). Western blot analysis showed decreased ABCC6 protein expression in liver of strains C3H and DBA as compared with HcB24 and B6 (Fig. 1B). Abcc6 genomic sequencing revealed 36 coding sequence variations in strains C3H and DBA compared with B6 (GenBank accession nos. EF109740 and EF 109741; SI Table 4). A 10-bp deletion in the 3′-UTR of the Abcc6 gene was found only in C3H and DBA and not in B6, B10 (SI Fig. 6), and several other inbred strains of mice resistant to DCC, including the A/J, MRL/MpJ, and BALB/cJ. The 3′-UTR can play an important role in translational control and mRNA stability, and mutations affecting the 3′-UTR have been shown to cause reduced level of transcript (14, 15). To test the possibility of this 10-bp deletion as the causal mutation, we performed a GFP reporter assay for chimeric constructs expressing 3′-UTR of the B6 or C3H allele under the control of CMV promoter (SI Fig. 7). No statistically significant difference was observed between the two chimeric constructs, although both demonstrated an inhibitory effect relative to the original construct. This suggests that the 10-bp deletion is not responsible for the altered expression although the anticipated regulatory effect could be liver cell-specific. RNA secondary structure predictions made by MFOLD program also suggest no significant differences in structural stability for the 3′ UTR of Abcc6 in B6 and C3H. We have also determined the 5′ flanking genomic sequence for the Abcc6 gene and found five sequence variations in the DBA strain (SI Fig. 8). Most of the putative promoter/enhancer elements predicted are not affected except one Sp1 site and two GC-boxes (16). A TATA-less promoter with Sp1 site-dependent translational regulatory mechanism has been proposed for Abcc6 gene expression (16).
Fig. 1.
Identification of Abcc6 as a candidate gene for the Dyscalc1 locus. (A) QPCR analysis of Abcc6 expression in liver, heart, and kidney (n = four mice each strain fed with chow diet; ∗, P < 0.05; †, P < 0.0001). Expression values for B6 homozygotes were set as 100. (B) Western blot analysis showing greatly reduced ABCC6 protein levels in liver in the DBA and C3H strains as compared with HcB24 and B6 strains.
Confirmation of Abcc6 as the Causal Gene for Dyscalc1.
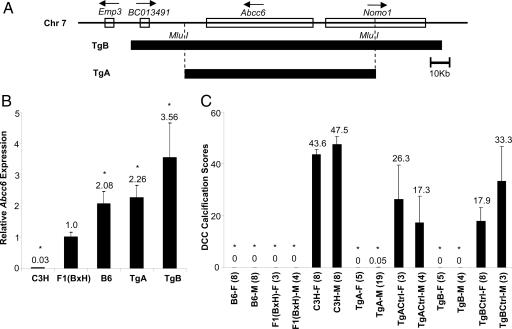
To validate Abcc6 as the causal gene, we used a transgenic complementation strategy. To ensure that the expression of the transgene occurs in the appropriate cell types and at the appropriate level, we used BAC clones containing the entire Abcc6 gene and flanking regions (at least 30-kb upstream and 10-kb downstream; Fig. 2A). The BAC library was constructed from strain B6 and the transgenes were bred onto a C3H genetic background. The transgenic TgA strain contains only the Abcc6 gene as the transgene, whereas the transgenic TgB strain contains Abcc6 and two additional genes. QPCR showed that in heterozygous transgenic mice, TgA and TgB, hepatic tissue expression levels of Abcc6 were equivalent to animals with two and three to four copies of the B6 functional allele, respectively (Fig. 2B). Phenotypic characterization showed that all heterozygous Abcc6 transgenic mice had 100% complementation of the DCC phenotype (i.e., little/no calcification despite a C3H background), whereas their nontransgenic littermates exhibited calcification (Fig. 2C). The mode of inheritance, as demonstrated by the F1 and the heterozygous transgenic mice, is consistent with a dominant mode for the B6 allele. Therefore, we conclude that Abcc6 is the underlying causal gene for Dyscalc1 and a major causal gene for DCC.
Fig. 2.
Confirmation of Abcc6 as the causal gene for Dyscalc1 using transgenic complementation strategy. (A) One B6-originated BAC clone was selected based on its coverage on chromosome 7 (closed boxes). The whole BAC, containing Abcc6 and two additional genes (BC013491 and Nomo1), was used to construct transgenic mice TgB. After restriction enzyme (Mlu I) digestion, the main fragment of the BAC containing only the Abcc6 gene was used to build transgenic mice TgA. (B) Confirmation of Abcc6 expression in transgenic mice. Hepatic tissue QPCR analysis of five strains are shown (n = four mice, each strain fed with HP diet for 4 weeks). Expression levels are normalized to the mean expression of Abcc6 observed in the F1 (BxH) mice. (C) DCC phenotypic characterization of all strains. Average values are indicated. F, female; M, male; Tg, transgenic; TgCtrl, nontransgenic littermate. The numbers of mice used for each group are in parentheses. ∗, P < 0.05.
Abcc6 Is Coregulated with Local Calcification Regulatory Genes and Genes from the BMP2-Wnt Pathway.
Towler and colleagues have recently proposed a major role for BMP2-Msx2-Wnt signaling pathway in arterial medial calcification using Msx2 transgenic mice (17). To assess the possible involvement of Abcc6 with this and other likely calcification mechanisms, we assembled a list of candidate genes for arterial calcification from the literature (18–20) and performed correlation analyses between their expression levels and that of Abcc6 using the BxD and BxH microarray data sets. In genetic crosses, transcript levels are subjected to hundreds of genetic perturbations, and genes in common pathways tend to exhibit coexpression (21). Therefore, by discovering robust correlations between genes with pertinent functions, we anticipated finding genes from closely related pathways. Of the 32 genes we tested for liver array data sets (SI Table 5), a total of seven genes were significantly correlated with Abcc6 in both crosses and six of them were directly associated with the local calcification regulators and the BMP2-Wnt pathway (SI Fig. 9).
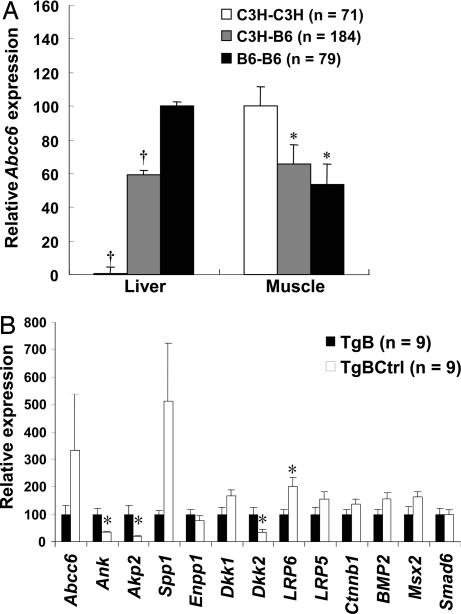
We also investigated the skeletal muscle gene expression profile, because dystrophic calcification after injury also occurs in skeletal muscle. By studying the gene expression variations in one of the afflicted tissues, we attempted to detect genes and pathways that contribute directly to calcium and phosphate regulation. We also included nine additional genes from the BMP2-Wnt signaling cascade, making the list a total of 41 genes. The expression level of Abcc6 in muscle is much lower than that in liver in B6 homozygote but interestingly was greater in the C3H homozygote than the heterozygote and the B6 homozygote (Fig. 3A). This Abcc6 expression level increase was confirmed using QPCR (chow diet treatment in SI Fig. 10, and HP diet treatment in Fig. 3B) in muscle of animals undergoing calcifying treatment, suggesting that Abcc6 expression is actively regulated in tissues that calcify. Using muscle data from the BxH intercross, a total of 34 of the 41 genes were found to significantly correlate with Abcc6 expression using either Pearson or Spearman correlation analysis (SI Table 6).
Fig. 3.
Abcc6 is coregulated with local calcification regulators and genes from the BMP2-Wnt pathway. (A) Abcc6 transcript levels in liver and muscle microarray data sets are significantly associated with Abcc6 allelic genotype, but in opposite directions. The BxH F2 mice were subdivided into three genotypic groups at the Abcc6 locus. Relative expression values were calculated from mean log ratio (“mlratio”; see SI Methods), and all values were adjusted to positive values. Expression of the homozygotes with the highest mlratio value was set as 100. (B) QPCR results for genes from skeletal muscle between the Abcc6 transgenic and littermate control mice fed with HP diet for 4 weeks. Relative expression values are compared with transcript levels in Abcc6 transgenic (100). ∗, P < 0.05; †, P < 0.0001.
These results suggest the involvement of multiple calcification pathways, including genes from the BMP2-Wnt pathway (e.g., Tnfα, LRP6, LRP5, Sfrp1, Wnt3a, Wnt5b, Wnt7a, Dkk1, Dkk2, Bmp2, Bmpr2, Smad6, Msx1, Msx2, and Sp7), the RANK/OPG pathway (e.g., Tnfsf11, Tnfrsf11a, and Tnfrsf11b), phosphate transporters and hydroxyapatite regulators (e.g., Slc20a1, Slc20a2, Enpp1, Akp2, Spp1, and ANK), and a major calcium homeostasis regulator (Cyp27b1). Three of the local calcification regulator genes (Ank, Akp2, and Enpp1) were among the top 10 genes significantly associated with Abcc6 (see SI Table 6).
To validate our findings in skeletal muscle, mRNA levels were measured by using QPCR, comparing Abcc6 transgenics and their nontransgenic littermates that had received a high-phosphate diet for 4 weeks (Fig. 3B). There was significantly decreased expression of Ank (3-fold) and Akp2 (5-fold) (P < 0.05) and increased expression of Enpp1 and Spp1 (1.5- and 5-fold, P = 0.12 and 0.17, respectively, using the Mann–Whitney test) in the nontransgenic control mice, accompanying the up-regulation of Abcc6 expression. There was ≈1.5 fold induction of Dkk1, LRP5, LRP6, Ctnnb1, BMP2, and Msx2 in the nontransgenic group, suggesting up-regulation of the BMP2-Wnt pathway, although only Dkk2 (3-fold decrease) and LRP6 (2-fold increase) reached statistical significance (P < 0.05). It should be noted that the power for detection of expression differences between control and transgenic mice is substantially less than for detecting correlations between gene expressions in the F2 mice because of the much smaller sample sizes in the former.
Expression of Abcc6 Is Associated with Decreased Systemic Osteopontin (OPN) Response.
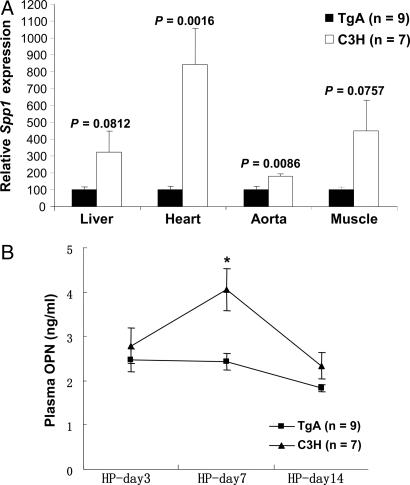
OPN, encoded by Spp1, is a major inducible local inhibitor of ectopic calcification that has been associated with Wnt signaling (22). Our data show that Spp1 expression is significantly negatively correlated with that of Abcc6 in both the liver and skeletal muscle microarray data sets (SI Tables 5 and 6). QPCR confirmed increased expression of Spp1 in liver, heart, aorta, and skeletal muscle tissues in Abcc6-deficient mice upon HP diet treatment (Fig. 4A). Interestingly, a locus on mouse chromosome 7 has been found to up-regulate Spp1 expression upon freeze–thaw injury in C3H mice associated with a plasma OPN surge 3 days after injury (23). In light of our finding, we speculated that this locus may correspond to Dyscalc1, and that Abcc6 may be the underlying causal gene. We compared the Abcc6 transgenic with the C3H control mice upon treatment with the HP diet (Fig. 4B) and observed a plasma OPN surge that appeared at day 7 for C3H mice, which was completely abolished in the transgenics. The temporal delay relative to the time of the OPN surge reported for the freeze–thaw injury model was possibly because of the HP diet being a relatively mild stimulus compared with the freeze–thaw injury for initiation of calcification.
Fig. 4.
Expression of Abcc6 is associated with decreased systemic OPN response. (A) QPCR results showed after HP diet treatment that there is an up-regulation of Spp1 gene in liver, heart, aorta, and muscle tissues in the C3H mice but not in Abcc6 transgenic mice. Relative expression values are compared with transcript levels in Abcc6 transgenics (100). P values are indicated. (B) A plasma OPN surge was observed for C3H mice after the mice were fed an HP diet for 7 days. The surge was completely abolished in Abcc6 transgenic mice. ∗, P < 0.05.
Discussion
DCC is a complex cardiovascular disorder influenced by multiple genetic as well as environmental factors. Using a traditional positional cloning strategy, we fine-mapped the major locus controlling the trait, Dyscalc1, to an 840-kb interval on chromosome 7. A cis-eQTL analysis was applied, and Abcc6 was identified as the primary candidate gene. BAC-mediated transgenic mice were generated and exhibited complete rescue of the calcification phenotype in C3H mice, confirming that Abcc6 is the underlying causal gene for Dyscalc1. We further explored the DCC disease mechanism with an integrative genomics approach by connecting Abcc6 expression with other genes and pathways known to be involved in systemic regulation of calcification, thereby providing a framework for future exploration of the disease mechanism.
Abcc6 is a member of a family of cellular export transporters and is predominantly expressed in liver and kidney (24). To date, ≈50 mutations of Abcc6 have been found in humans with the autosomal recessive disease, pseudoxanthoma elasticum (PXE), manifested by dystrophic calcification affecting the elastic fibers in skin, Bruch's membrane, and vessel walls with highly variable clinical manifestations (25). It is noteworthy that myocardial calcification has not been reported as a phenotype associated with either human PXE or the mouse Abcc6 knockout models (26, 27). Moreover, Abcc6 knockout mice on a B6 background develop medial vascular calcification spontaneously with age, whereas the Abcc6-deficient C3H mice do not show signs of vascular calcification until challenged by hyperlipidemia (unpublished data). These differences could result from the different genetic backgrounds or from residual Abcc6 expression in mice with the C3H Dyscalc1 allele. In light of this, we propose that genetic backgrounds and/or varying levels of Abcc6 expression are underlying mechanisms for the highly variable clinical manifestations of PXE. Further studies using the two animal models should shed more light on the disease mechanism of PXE and DCC and of ABCC6 transporter function.
Abcc6 was found to have the highest expression in liver and kidney, although low levels of mRNA have been found in many tissues, including cardiac and skeletal muscle (28). Our studies indicate that Abcc6 is actively regulated in skeletal muscle tissue upon injury, implying that the transporter may have important local functions in injury responses or remodeling processes. Transcription analysis of the human Abcc6 promoter region identified sequences homologous for known TGF-β responsive Sp1 binding elements, and both TGF-β and TNF-α have been shown to stimulate human Abcc6 expression in vitro (29). It is tempting to speculate that the up-regulation of Abcc6 upon injury may be directly influenced by these cytokines.
Using an F2 segregating population, significant correlations of expression levels were observed between Abcc6 and genes from the local calcification regulatory system and known systemic calcification pathways such as the BMP2-Wnt pathway. Using Abcc6 transgenic and littermate control mice treated with an HP diet, QPCR analysis demonstrated significant expression changes for Ank, Akp2, Dkk2, and LRP6 and suggestive expression changes for Dkk1, LRP5, LRP6, Ctnnb1, BMP2, and Msx2, all key players in the BMP2-Wnt pathway, suggesting involvement of this pathway.
OPN is an acidic phosphoprotein found at high levels in calcified vascular tissues. Recent studies demonstrated OPN to be a potent inhibitor for mineralization both in vitro and in vivo (30). Normally expressed at low levels in blood vessels, heart, and skeletal muscles [Genomics Institute of the Novartis Research Foundation (GNF) SymAtlas: http://symatlas.gnf.org/SymAtlas], OPN is rapidly induced upon injury and presumably functions to block the deposition of hydroxyapatite and promote crystal resorption through activation of monocytes (30). Our data from F2 population correlation analysis, QPCR results, and plasma OPN ELISA data (especially data from liver, where it is the primary tissue for Abcc6 expression and also not affected by calcification) were all consistent with a strong association of Abcc6 expression with significantly decreased OPN expression upon initiation of calcification, suggestive of an inhibitory role for Abcc6 expression on Spp1 expression regulation.
The identification of genes contributing to complex traits is one of the major challenges in modern biology and medicine. Although loci contributing to these traits can be mapped in a straightforward manner in animal models, success in identifying the underlying genes has been achieved only rarely. A recent review indicated that, although >2,000 QTL have been mapped over the past 15 years, only 22 of the underlying genes have been identified (31). As demonstrated in the current study, the integrative genomics approach provides an approach for the identification of causal genes and related pathways. However, as for any method, there are limitations for this application. First, not all traits can be analyzed by using this approach, because not all traits involve differences in gene transcript abundance. Second, an appropriate tissue has to be available for microarray expression analysis, although as pointed out earlier, cis-variations frequently can be detected in multiple tissues. Finally, different experimental conditions may have significantly different impacts on the outcome of the pathway analysis. Nevertheless, as databases of expression data grow, our approach promises to be a valuable complement in gene identification and to an understanding of the related pathways (32).
Materials and Methods
Animals.
Inbred strains of C57BL/6J, C57BL/10J, DBA/2J, C3H/HeJ, A/J, MRL/MpJ, and BALB/cJ mice were obtained from The Jackson Laboratory (Bar Harbor, ME). The RC mice were originally developed by Demant and Hart (9) from strains B10 and C3H/DiSnA (HcB C3H). All animals were maintained in specific pathogen-free facilities meeting the guidelines of the Association for Accreditation of Laboratory Animal Care. RC HcB24 strain was backcrossed with C3H mice to produce N2 mice. Those with the desired genotypes were placed on an HP diet containing calcium (0.4%), phosphate (0.85%), and low (0.04%) magnesium (Teklad TD00442, Harlan-Teklad, Madison, WI) for phenotypic characterization (see SI Methods).
Genotyping and Sequencing.
Genomic sequence for C57BL/6J (build 34) was obtained from Ensembl (www.ensembl.org) and the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) web sites. Gene Runner software (Hastings Software, Hastings, NY) was used for primer design. DNA was isolated from ear or tail tissues by using DNeasy Tissue Kit (Qiagen, Valencia, CA). PCR products for microsatellite markers were resolved by agarose gel electrophoresis or automated ABI 3700 sequencers (Applied Biosystems, Foster City, CA). Lasergene, version 5.0, from DNAStar (Madison, WI), was used for sequence editing, assembly, and alignment of multiple sequences. Prediction of RNA secondary structure was performed by using the mFOLD program (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html).
The 31 exons of the Abcc6 gene were amplified by using genomic DNA and reverse-transcribed kidney cDNA from the C3H and DBA strains. For other inbred strains, including C57BL/6J, C57BL/10J, A/J, MRL/MpJ, and BALB/cJ, genomic DNA was used in screening for the 10-bp deletion in the 3′-UTR. The PCR product was amplified by using blunt-ending Pfu polymerase (Invitrogen, Carlsbad, CA), cloned into the pCR-Blunt II-TOPO vector (Invitrogen), miniprepped with Qiagen Miniprep Kit (Qiagen), and sequenced by using Big Dye Terminator cycle sequencing (Qiagen) and automated ABI 3700 sequencers (Applied Biosystems).
RNA Isolation and QPCR.
Freshly isolated tissues were used for RNA isolation by using TRIzol reagent (Invitrogen), followed by an RNeasy Mini Kit (Qiagen) cleanup step. Complementary DNA was synthesized with SuperScript III First-Strand Synthesis Systems for RT-PCR (Invitrogen). QPCR was carried out by using QuantiTect SYBR Green PCR Kit (Qiagen) and the ABI prism 7000 Sequence Detection System (Applied Biosystems). The primers were designed to bridge one splice site, and the product was sequenced to confirm that the correct gene was amplified. Several pairs of QPCR primers were obtained from the Primer Bank web site (http://pga.mgh.harvard.edu/primerbank) (33) or public literature (34). If not specified, expression levels were normalized to that of the housekeeping gene β-actin. Each QPCR was carried out at least in duplicate.
Western Blot.
Total hepatic protein extract was prepared from frozen liver tissues. Briefly, a 100-mg sample of liver tissue was rinsed with PBS and homogenized in 400 μl of extraction buffer containing 50 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% Na-deoxycholate, and 0.1% SDS. After centrifugation, 60 μg of total protein extract was resolved by SDS/PAGE by using 10% NuPAGE-Novex Bis-Tris Gel (Invitrogen), then electrotransferred to Hybond-P PVDF membrane (Amersham, Piscataway, NJ). K14 primary antibody for Abcc6 was the kind gift of Bruno Stieger at University Hospital, Zurich, Switzerland. Primary antibody for β-actin was purchased from Sigma (St. Louis, MO). ECL Western Blotting Detection Reagents and analysis system (Amersham) and KODAK BioMax Light Film (Kodak, Rochester, NY) were used for signal detection.
Construction of Transgenic Mice.
BAC RP24–179C21 was selected based on BAC contig information from University of California, Santa Cruz, Genome Bioinformatics web server (http://genome.ucsc.edu) and purchased from BAC PAC Resource at Children's Hospital Oakland Research Institute (Oakland, CA). Intact BAC DNA was extracted by Qiagen Maxi-prep Kit (Qiagen) and pulse-field gel electrophoresis, followed by Sepharose column purification as described (University of Michigan Transgenic Animal Model Core, University of Michigan Medical School, Ann Arbor, MI). Mlu I (New England Biolabs, Ipswich, MA) restriction digestion was used to remove the vector sequence as well as the two additional genes BC013491 and Nomo1 from the BAC. Abcc6 transgenic mice were produced by DNA microinjection using F2(BxH) fertilized eggs. The service was provided by the Transgenic Mouse Facility of the University of California, Irvine, CA. Transgenic founder mice were backcrossed to C3H strain, and their progeny were selected for animals containing the Abcc6 transgene. During this selective breeding process, progeny were selected for C3H homozygous allele at all four Dyscalc loci.
Histological Analyses.
For DCC phenotypic characterizations, starting at 6–8 weeks of age, age-matched mice were placed on an HP diet. After 4 weeks, the heart tissue was collected as described (8). Methods for determining arterial calcification, aortic aneurysms, and atherosclerotic lesions, were described (35, 36). For further information, see SI Methods.
OPN Plasma Concentration Analysis.
To determine whether plasma OPN level response to the HP diet is regulated by the Abcc6 gene, transgenic mice and a C3H control were treated with an HP diet starting from 8 weeks of age. Plasma OPN levels were determined at specified time points (at days 3, 7, and 14 after the start of the HP diet) using the ELISA-based mouse OPN assay kit (Catalogue 900-090; Assay Designs, Inc., Ann Arbor, MI).
Statistical Analyses.
Detailed information about the BxD and BxH intercrosses has been reported (10, 11). For further information on microarray analysis, see SI Methods. Student's t test or ANOVA statistical analyses were carried out by using the StatView software (SAS Institute, Cary, NC). The nonparametric Wilcoxon Mann–Whitney test and Pearson and Spearman correlation analyses were performed by using the SAS program (SAS Institute). A significance threshold of P < 0.05 was used. Bonferroni correction was used for all types of multiple analyses.
Supplementary Material
Acknowledgments
We thank Bruno Stieger (University Hospital, Zurich, Switzerland) for sharing his ABCC6 antibody (K14), Sudheer Doss for suggestions about the statistical analyses of microarray databases, and Richard Davis for reviewing the manuscript. This work was supported by the National Institutes of Health and the American Heart Association [Grants P01 HL30568 (to A.J.L. and T.A.D.) and 0425134Y (to H.M.)].
Abbreviations
- DCC
dystrophic cardiac calcification
- HP
high phosphorous
- QTL
quantitative trait locus
- eQTL
expression QTL
- RC
recombinant congenic
- QPCR
quantitative RT-PCR
- OPN
osteopontin
- PXE
pseudoxanthoma elasticum
- BxH
intercross between B6 and C3H mice
- BxD
intercross of B6 and DBA mice.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607620104/DC1.
References
- 1.Brean HP, Marks JH, Sosman MC, Schlesinger MJ. Radiology. 1950;54:33–42. doi: 10.1148/54.1.33. [DOI] [PubMed] [Google Scholar]
- 2.Brunnert SR. Lab Anim Science. 1997;47:11–18. [PubMed] [Google Scholar]
- 3.Van den Broek FA, Beynen AC. Lab Anim. 1998;32:483–491. doi: 10.1258/002367798780599758. [DOI] [PubMed] [Google Scholar]
- 4.Korff S, Riechert N, Schoensiegel F, Weichenhan D, Autschbach F, Katus HA, Ivandic BT. Virchows Arch. 2005;448:630–638. doi: 10.1007/s00428-005-0071-7. [DOI] [PubMed] [Google Scholar]
- 5.Van den Broek FA, Bakker R, den Bieman M, Fielmich-Bouwman AX, Lemmens AG, van Lith HA, Nissen I, Ritskes-Hoitinga JM, van Tintelen G, van Zutphen LF. Biochem Biophy Res Commun. 1998;253:204–208. doi: 10.1006/bbrc.1998.9776. [DOI] [PubMed] [Google Scholar]
- 6.Ivandic BT, Utz HF, Kaczmarek PM, Aherrahrou Z, Axtner SB, Klepsch C, Lusis AJ, Katus HA. Physiol Genom. 2001;6:137–144. doi: 10.1152/physiolgenomics.2001.6.3.137. [DOI] [PubMed] [Google Scholar]
- 7.Ivandic BT, Qiao J, Machleder D, Liao F, Drake TA, Lusis AJ. Proc Natl Acad Sci USA. 1996;93:5483–5488. doi: 10.1073/pnas.93.11.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colinayo VV, Qiao J, Demant P, Krass K, Lusis AJ, Drake TA. Mamm Genome. 2002;13:283–288. doi: 10.1007/s00335-001-2148-1. [DOI] [PubMed] [Google Scholar]
- 9.Demant P, Hart AAM. Immunogenetics. 1986;24:416–422. doi: 10.1007/BF00377961. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Yehya N, Schadt EE, Wang H, Drake TA, Lusis AJ. PLoS Genet. 2006;2:e15. doi: 10.1371/journal.pgen.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Miligan SB, Lamb JR, Cavet G, et al. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doss S, Schadt EE, Drake TA, Lusis AJ. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glazier AM, Scott J, Aitman TJ. Mamm Genome. 2002;13:108–113. doi: 10.1007/s00335-001-2132-9. [DOI] [PubMed] [Google Scholar]
- 15.Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AF. Nat Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- 16.Douet V, VanWart CM, Heller MB, Reinhard S, Le Saux O. Biochim Biophys Acta. 2006;1759:426–436. doi: 10.1016/j.bbaexp.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao JS, Cheng SL, Pingsterhaus JM, Charlton-Kachigian N, Loewy AP, Towler DA. J Clin Invest. 2005;115:1210–1220. doi: 10.1172/JCI24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wozney JM, Rosen V. Clin Orthop Relat Res. 1998:26–37. [PubMed] [Google Scholar]
- 19.Giachelli C. J Am Soc Nephrol. 2004;15:2959–2964. doi: 10.1097/01.ASN.0000145894.57533.C4. [DOI] [PubMed] [Google Scholar]
- 20.Wallin R, Wajih N, Greenwood GT, Sane DC. Med Res Rev. 2001;21:274–301. doi: 10.1002/med.1010. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Burmeister M. Hum Mol Genet. 2005;14:R163–R169. doi: 10.1093/hmg/ddi267. [DOI] [PubMed] [Google Scholar]
- 22.Vattikuti R, Towler DA. Am J Physiol. 2004;286:E686–E696. doi: 10.1152/ajpendo.00552.2003. [DOI] [PubMed] [Google Scholar]
- 23.Aherrahrou Z, Axtner SB, Kaczmarek PM, Jurat A, Korff S, Doehring LC, Weichenhan D, Katus HA, Ivandic BT. Am J Pathol. 2004;164:1379–1387. doi: 10.1016/S0002-9440(10)63224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madon J, Hagenbuch B, Landmann L, Meier PJ, Stieger B. Mol Pharmacol. 2000;57:634–641. doi: 10.1124/mol.57.3.634. [DOI] [PubMed] [Google Scholar]
- 25.Le Saux O, Urban Z, Tschuch C, Csiszar K, Bacchelli B, Quaglino D, Pasquali-Ronchetti I, Pope FM, Richards A, Terry S, et al. Nat Genet. 2000;25:223–227. doi: 10.1038/76102. [DOI] [PubMed] [Google Scholar]
- 26.Gorgels TG, Hu X, Scheffer GL, van der Wal AC, Toonstra J, de Jong PT, van Kuppevelt TH, Levelt CN, de Wolf A, Loves WJ, et al. Hum Mol Genet. 2005;14:1763–1773. doi: 10.1093/hmg/ddi183. [DOI] [PubMed] [Google Scholar]
- 27.Klement JF, Matsuzaki Y, Jiang QJ, Terlizzi J, Choi HY, Fujimoto N, Li K, Pulkkinen L, Birk DE, Sundberg JP, Uitto J. Mol Cell Biol. 2005;25:1–12. doi: 10.1128/MCB.25.18.8299-8310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beck K, Hayashi K, Nishiguchi B, Le Saux O, Hayashi M, Boyd CD. J Histochem Cytochem. 2003;51:887–902. doi: 10.1177/002215540305100704. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Q, Matsuzaki Y, Li K, Uitto J. J Invest Dermatol. 2006;126:325–335. doi: 10.1038/sj.jid.5700065. [DOI] [PubMed] [Google Scholar]
- 30.Steitz SA, Speer MY, Curinga G, Yang H, Haynes P, Aeberosold R, Schinke T, Karsenty G, Giachelli CM. Am J Pathol. 2002;161:2035–2046. [Google Scholar]
- 31.Flint J, Valdar W, Shifman S, Mott R. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 32.Giallourakis C, Henson C, Reich M, Xie X, Mootha VK. Annu Rev Genom Hum Genet. 2005;6:381–406. doi: 10.1146/annurev.genom.6.080604.162234. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Seed B. Nucleic Acids Res. 2003;31:1–8. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng SL, Shao JS, Charlton-Kachigian N, Loewy AP, Towler DA. J Biol Chem. 2003;278:45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 35.Qiao JH, Xie PZ, Fishbein MC, Kreuzer J, Drake TA, Demer LL, Lusis AJ. Arterioscler Thromb Vasc Biol. 1994;14:1480–1497. doi: 10.1161/01.atv.14.9.1480. [DOI] [PubMed] [Google Scholar]
- 36.Ghazalpour A, Wang X, Lusis AJ, Mehrabian M. Genetics. 2006;173:943–951. doi: 10.1534/genetics.106.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.