Abstract
In plants, as in most eukaryotic cells, import of nuclear-encoded cytosolic tRNAs is an essential process for mitochondrial biogenesis. Despite its broad occurrence, the mechanisms governing RNA transport into mitochondria are far less understood than protein import. This article demonstrates by Northwestern and gel-shift experiments that the plant mitochondrial voltage-dependent anion channel (VDAC) protein interacts with tRNA in vitro. It shows also that this porin, known to play a key role in metabolite transport, is a major component of the channel involved in the tRNA translocation step through the plant mitochondrial outer membrane, as supported by inhibition of tRNA import into isolated mitochondria by VDAC antibodies and Ruthenium red. However VDAC is not a tRNA receptor on the outer membrane. Rather, two major components from the TOM (translocase of the outer mitochondrial membrane) complex, namely TOM20 and TOM40, are important for tRNA binding at the surface of mitochondria, suggesting that they are also involved in tRNA import. Finally, we show that proteins and tRNAs are translocated into plant mitochondria by different pathways. Together, these findings identify unexpected components of the tRNA import machinery and suggest that the plant tRNA import pathway has evolved by recruiting multifunctional proteins.
Keywords: mitochondrial porin, cytosolic tRNAs, import factor
Mitochondrial genomes vary in the number of expressed tRNA genes, ranging from none, as in some kinetoplastid protozoans, to the complete set required for mitochondrial protein synthesis, as in humans. Other organisms fall between these two extremes; for example, land plant mitochondrial genomes lack at least one-third of the tRNA genes. As a consequence, most eukaryotic cells had to evolve mechanisms for delivering nuclear-encoded tRNAs from the cytosol into mitochondria.
To date, tRNA import into mitochondria has been studied mainly in the yeast Saccharomyces cerevisiae, in protozoans like Trypanosoma or Leishmania, and in higher plants (1, 2). In agreement with a polyphyletic origin of RNA import into mitochondria, the data obtained so far led to different mechanistic models. Import of tRNALys(CUU) into S. cerevisiae mitochondria was the first to be studied in detail. Surprisingly, it was recently shown that the aminoacylated form of this tRNA is first specifically recognized in the cytosol by the glycolytic enzyme enolase, which addresses it toward the mitochondrial surface. The tRNA is then transferred to the perimitochondrially synthesized precursor form of mitochondrial LysRS before its targeting to the mitochondrial matrix through the outer membrane preprotein import complex (3). By contrast, the ability of two uncharged tRNAGln isoacceptors to be imported in vitro into S. cerevisiae mitochondria in the absence of added cytosolic factors supports the idea that in this case the mechanism differs radically from that of tRNALys import (4). In trypanosomatids, as found mainly in Trypanosoma and Leishmania, mitochondrial tRNA import does not require cytosolic proteins, but involves protease-sensitive outer membrane receptors and is ATP-dependent. Furthermore, data support different mechanisms for tRNA and protein import but lead also to conflicting conclusions. In particular, the requirement of a membrane potential was attributed either for translocation of synthetic transcripts in Leishmania donovanii and Leishmania tropica or for import of tandemly linked tRNAs in Trypanosoma brucei (5, 6). By contrast, dissipation of the membrane potential by the ionophore valinomycin does not abolish import of tRNA fragments in T. brucei and mature tRNA transcripts in Leishmania tarentolae (7, 8). Likewise import determinants in trypanosomatid tRNAs remain controversial (1, 2, 9). On the other hand, information on the components of the tRNA import apparatus remains scarce. A 15-kDa putative import tRNA receptor has been reported in L. tropica, but this protein remains to be identified (10). More recently, it has been shown in L. tropica that a multisubunit RNA import complex (RIC) located on the inner mitochondrial membrane is implicated in tRNA import (11) and two subunits, RIC1 (a structural homologue to the α subunit of F1 ATP synthase) and RIC8 (an homologue to subunit 6b of ubiquinol cytochrome c reductase) were identified (12, 13). However, considering the contradictory data obtained so far, a deeper understanding of the import factors involved in different trypanosomatids will be important in the future.
In plants, recent developments showed that tRNA import is an ATP-dependent process, does not require any added cytosolic factors, and includes at least one protease-sensitive component on the surface of mitochondria (14). Plant mitochondrial tRNA import can be inhibited in vitro by valinomycin or oligomycin, meaning that a membrane potential and a functional respiratory chain are required. As a step toward understanding plant tRNA import, it is now essential to better dissect the protein factors implicated at the level of the mitochondrial membranes. Here we demonstrate that the voltage-dependent anion channel (VDAC), known to play a major role in the transport of metabolites, is a key component of the channel involved in the tRNA translocation step through the plant mitochondrial outer membrane. Our data also suggest that TOM20 and TOM40, two major components of the protein translocase of the outer mitochondrial membrane (TOM) complex, are implicated in the binding of tRNAs on the surface of mitochondria. Thus they play an essential role not only in protein import but also in tRNA import. Finally, we provide evidence that proteins and tRNAs are imported into plant mitochondria via different pathways. As a whole, our findings bring an additional view of the evolution of plant tRNA import machinery by recruiting multifunctional proteins.
Results
Potato Mitochondrial VDAC Interacts with tRNA in Vitro.
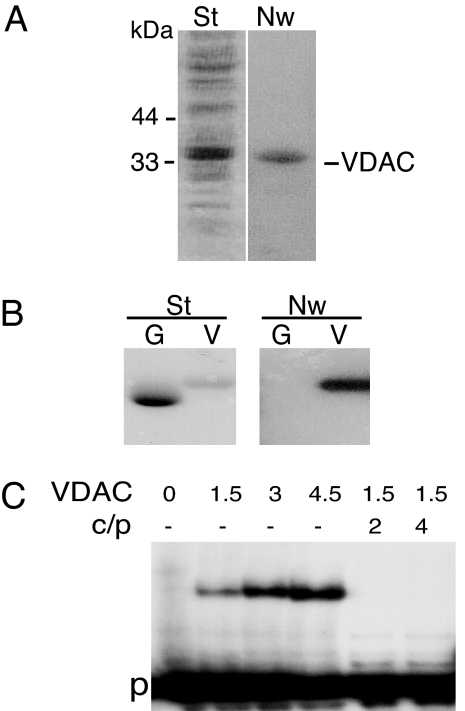
Total proteins purified from Solanum tuberosum outer mitochondrial membranes were used to perform a Northwestern experiment in the presence of radiolabeled plant cytosolic tRNAAla. A strong signal was obtained with a protein migrating at 34 kDa (Fig. 1A). The purified protein was identified by N-terminal sequencing (KGPGLYTEIGKKAxDLLY with x being an unidentified amino acid) as VDAC of potato mitochondrial outer membrane, a protein previously characterized (15). To confirm that the VDAC protein was able to interact with tRNA, the protein was overexpressed in Escherichia coli and purified by His-tag affinity. As shown by Northwestern experiments (Fig. 1B), whereas no signal was observed with overexpressed GFP, overexpressed VDAC strongly interacted with labeled tRNAAla transcript. This interaction was supported by gel-shift assay performed with labeled tRNAAla transcript as a probe (Fig. 1C). In the presence of increasing concentrations of overexpressed VDAC, increasing amounts of tRNA–VDAC complex were detected, whereas the addition of an excess of unlabeled tRNAAla transcripts completely blocked by competition binding between labeled tRNAAla transcript and VDAC.
Fig. 1.
S. tuberosum mitochondrial VDAC interacts with tRNA in vitro. (A) S. tuberosum mitochondrial proteins from outer membrane after SDS/PAGE fractionation, transfer onto nylon membrane, and staining with Coomassie blue (St). For Northwestern blot analysis, the membrane, after protein renaturation, was incubated with labeled in vitro-transcribed cytosolic A. thaliana tRNAAla. After incubation and washing, the blot was subjected to autoradiography (Nw). Molecular masses of marker proteins are indicated. (B) Coomassie blue-stained profile (St) and Northwestern analysis (Nw) of His-tagged purified GFP (G) and VDAC (V) proteins. (C) Gel-shift assay performed with labeled tRNAAla as a probe (p) in the presence of increasing amounts (in picomoles) of purified VDAC protein. For competition assays, 2- to 4-fold excess of unlabeled tRNAAla (c) was included in the reaction mixture, and the ratio competitor/probe (c/p) is indicated.
In Vitro tRNA Import into Isolated Mitochondria Is Inhibited by VDAC Antibodies and Ruthenium Red (RuR).
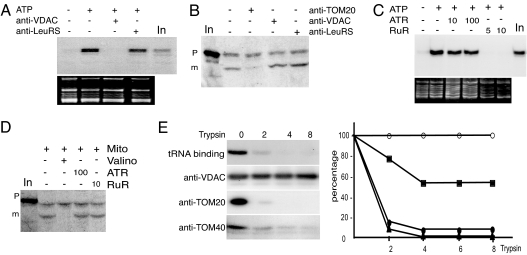
The involvement of VDAC in mitochondrial tRNA import was examined by testing the effect of potato mitochondrial VDAC antibodies on in vitro tRNAAla import into isolated mitochondria. As previously shown, tRNA import is a physiological ATP-dependent process (14). Thus, as a control, all in vitro assays presented here were performed with and without ATP, and the control with ATP was taken as reference (Figs. 2–4). As reported (14) and on the average, the amount of RNase protected transcript when import was carried out in the presence of ATP fluctuates between 0.2% and 0.5% of the input. As shown in Fig. 2A, tRNA uptake was completely abolished when mitochondria were pretreated with specific anti-VDAC antibodies before the import assay (for the specificity of antibodies see Fig. 5A, which is published as supporting information on the PNAS web site). In contrast, no inhibition (Figs. 2A and 3A) was observed with either mock-treated mitochondria or mitochondria pretreated with antibodies against plant cytosolic LeuRS or subunit 9 of the inner mitochondrial membrane complex I (NAD9). The tRNAAla substrate was not significantly degraded by the different antisera (data not shown). Thus, the lack of tRNA incorporation into isolated mitochondria pretreated with VDAC antibodies was not caused by tRNA degradation during the assays. As VDAC is the major component of the mitochondrial outer membrane, we checked the possibility that VDAC antibodies could inhibit tRNA import unspecifically by saturating the outer membrane. To exclude this hypothesis, we performed in parallel in vitro import of the fusion protein GluRS-GFP (16) into isolated mitochondria (Fig. 2B). For this protein, import efficiency into isolated S. tuberosum mitochondria was ≈5% of the input. Antibodies against LeuRS used as control and against VDAC had no effect on GluRS-GFP import into isolated potato mitochondria. As expected, an antiserum raised against TOM20, the mitochondrial receptor of the protein import channel, inhibited ≈75% of the uptake of GluRS-GFP into mitochondria (average of three independent experiments).
Fig. 2.
Implication of VDAC in mitochondrial tRNA import. (A and C) In vitro import of tRNA into isolated mitochondria. Labeled in vitro-transcribed tRNAAla (105 cpm) was incubated with isolated mitochondria in the absence (−) or presence (+) of 5 μl of antibodies against VDAC or LeuRS (A) and 5 or 10 μM RuR or 10 or 100 μM ATR (C). Incubations performed in the absence (−) or presence (+) of 5 mM ATP were used as control experiments. In, input RNAs (100 cpm). The corresponding ethidium bromide-stained gels are shown below the autoradiograms. Incubation of tRNAAla transcript with mitochondria in the presence of increasing concentration of VDAC antibodies shows that 1 μl of VDAC antibodies completely inhibits import (data not shown). (B and D) In vitro import of protein into isolated mitochondria. Labeled in vitro-synthesized GluRS targeting sequence fused to GFP was incubated with mitochondria in the absence (−) or presence (+) of 5 μl of antibodies against TOM20, VDAC, or LeuRS (B) or 10 μM RuR or 100 μM ATR (D). Upon import into mitochondria, the targeting presequence is cleaved off from the precursor protein (p), and a smaller protein corresponding to mature form (m) appears. As a control, protein import was shown to depend on mitochondrial membrane potential, as demonstrated by inhibition of import by valinomycin. In, input radiolabeled protein GluRS-GFP. (E) Binding of tRNAAla to isolated mitochondria depends on trypsin-sensitive proteins. To remove proteins exposed on the outer membrane, isolated mitochondria were treated with increasing concentrations of trypsin (0, 2, 4, and 8 μg of trypsin per mg of mitochondrial proteins) as described (14). Binding was performed under the same conditions as for tRNA import except that the RNase digestion step was omitted after incubation with labeled tRNAAla. As previously reported, the amount of bound tRNAs onto mitochondria is ≈2–5% of the input (compared with 0.2–0.5% of the input for RNase-protected imported tRNAs). Corresponding mitochondrial protein samples were analyzed by Western blot, using antibodies directed against VDAC, TOM20, and TOM40. For graphical representation of the results, the amounts of bound tRNAs (●), TOM20 (▴), TOM40 (■), and VDAC (○) were quantified with MacBAS 100 software.
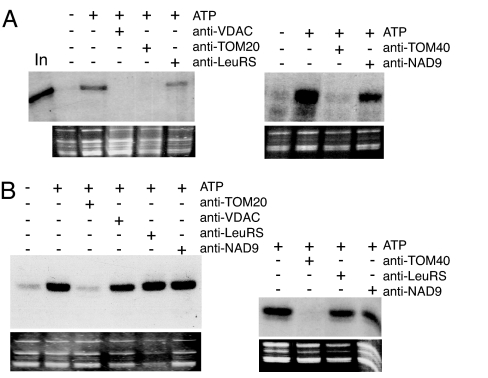
Fig. 3.
Both tRNA import and tRNA binding are inhibited by TOM20 and TOM40 antibodies. (A) In vitro import of tRNA into isolated mitochondria. Labeled in vitro-transcribed tRNAAla (105 cpm) was incubated with isolated mitochondria in the absence (−) or presence (+) of 5 μl of antibodies raised against VDAC, TOM20, TOM40, LeuRS, or NAD9. The corresponding ethidium bromide-stained gels are shown below the autoradiograms. In, input RNAs (1,000 cpm). Incubation of tRNAAla transcript with mitochondria in the presence of increasing concentration of TOM20 and TOM40 antibodies shows that 1 μl of the antibodies completely inhibits import (data not shown). (B) Binding of tRNA to isolated mitochondria. Binding was performed as described (14). Incubations were performed in the absence (−) or presence (+) of 5 mM ATP. Note that between A and B, exposures times for the autoradiography are very different (≈10 times longer exposure for import than for binding).
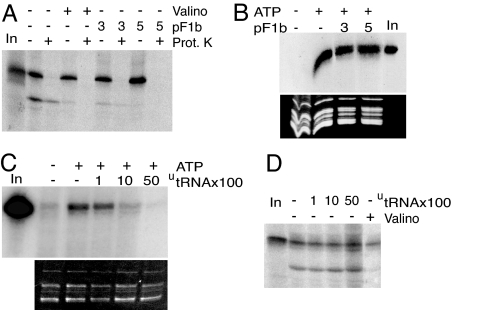
Fig. 4.
F1β presequence competes for protein import but not for tRNA import, and unlabeled tRNA competes for tRNA import but not for protein import. (A) Labeled in vitro-synthesized GluRS-GFP protein was incubated with isolated mitochondria in the absence (−) or presence of 3 or 5 μM F1β presequence (pF1b). After incubation, samples were treated (+) or not (−) with proteinase K (Prot. K). As a control, protein import was inhibited in the presence (+) of valinomycin (Valino). In, input-labeled protein GluRS-GFP. (B) Labeled in vitro-transcribed tRNAAla (105 cpm) was incubated with isolated mitochondria in the absence (−) or presence of 3 or 5 μM F1β presequence. As a control, incubations were performed in the absence (−) or presence (+) of 5 mM ATP. The corresponding ethidium bromide-stained gel is shown below the autoradiogram. In, input RNAs (100 cpm). (C) Competition experiments of tRNAAla import into isolated mitochondria using unlabeled tRNAAla as competitor (utRNA). The experiments were performed in the absence (−) or presence (+) of 5 mM ATP. In, input RNAs (1,000 cpm). (D) Competition experiments of GluRS-GFP protein import into isolated mitochondria using unlabeled tRNAAla as competitor (utRNA). As a control, protein import was run in the absence (−) or presence (+) of valinomycin (Valino). In, input-labeled protein GluRS-GFP.
In another approach, we took advantage of the fact that RuR has been previously shown to induce the closure of VDAC in rat liver mitochondria (17). We thus tested its effect on tRNA import. As shown in Fig. 2C, complete inhibition of tRNA import was observed when mitochondria were pretreated with RuR, whereas GluRS-GFP import was not affected by the addition of RuR (Fig. 2D). VDAC represents the main pathway for ATP exchange through the mitochondrial outer membrane, and the abolition of tRNA import could have been a consequence of this ATP exchange inhibition. However, when mitochondria were pretreated with atractyloside (ATR), an inhibitor of the ADP/ATP translocator (ANT) (18), tRNA import was not affected (Fig. 2C). Thus perturbing the overall ADP/ATP exchange at the level of mitochondrial membranes does not affect tRNA import. Alternatively, as protein import is affected neither by RuR nor by ATR (Fig. 2D), ATP provided externally and by respiratory chain activity is sufficient without need of a functional ADP/ATP exchange via VDAC or ANT. Altogether, the inhibition of in vitro tRNA import obtained by two independent means, VDAC antibodies and RuR, demonstrates that VDAC is involved in tRNA import into potato mitochondria.
We previously showed that trypsin treatment of mitochondria before in vitro assay completely abolishes tRNAAla import (14). We now show that trypsin-treated mitochondria also lose their ability to bind labeled tRNAAla transcript (Fig. 2E). Both data suggest the presence of at least one protease-sensitive outer membrane tRNA receptor at the surface of mitochondria. In contrast, Western blot analysis indicates that VDAC is fully resistant to this protease (Fig. 2E). This finding suggests that VDAC cannot be the mitochondrial outer membrane tRNA receptor but rather is a translocator.
TOM Proteins Participate in tRNA Binding and Are Part of the tRNA Import Channel.
Potential candidates for outer membrane tRNA receptors are proteins belonging to the TOM complex. Therefore we tested the effect of antibodies raised against TOM20, the mitochondrial receptor for protein import, and TOM40, a protein-conducting channel (19). Incubation of mitochondria with TOM20- or TOM40-specific antibodies (for the specificity antibodies, see Fig. 5A), led to, respectively, 75% and 100% inhibition not only of GluRS-GFP import (Figs. 2B and 5B) but also of tRNA import (Fig. 3A). Furthermore, in contrast to VDAC, TOM20 was fully sensitive to trypsin, and its degradation profile is similar to the loss of tRNA binding, whereas the degradation of TOM40 is slower and only partial (Fig. 2E).
To investigate whether TOM proteins might play the role of tRNA receptors at the surface of mitochondria, purified mitochondria were tested for their ability to fix tRNAAla transcript in vitro in the presence of various antibodies. As shown in Fig. 3B, binding of tRNAAla transcript to the outer membrane is completely abolished when mitochondria were preincubated with either TOM20 or TOM40 antibodies. As controls, no inhibition of tRNA binding was observed with mock-treated mitochondria or mitochondria preincubated with antibodies against LeuRS or NAD9. In addition, a tRNA binding signal comparable to those obtained in control experiments was observed in the presence of VDAC antibodies, further demonstrating that VDAC is not the outer membrane receptor. Furthermore, it is worth remembering that Northwestern experiments (Fig. 1A) detected a strong interaction between tRNAs and VDAC but no interaction with either TOM20 or TOM40. By contrast, VDAC antibodies do not inhibit tRNA binding onto intact mitochondria, whereas TOM20 or TOM40 antibodies do. This apparent discrepancy can be rationalized. First, as shown in Fig. 1, VDAC is ≈50-fold more abundant than TOM20 in plant mitochondrial outer membrane (15, 20). It is thus not surprising, because Northwestern is not a technique sensitive enough, that only VDAC was identified by this approach. Furthermore, this technique is based on the ability of proteins to renature and the possibility exists that TOM proteins were not properly refolded under our experimental conditions. Finally, one cannot rule out that the binding of tRNAs at the surface of mitochondria requires the coordinated action of different proteins, and that TOM20 or TOM40 taken separately cannot interact efficiently with tRNAs. Taken together, this result supports the idea that binding of tRNAs to the mitochondrial outer membrane requires the concerted action of several proteins and, in particular, at least two components of the TOM complex, TOM20 and TOM40.
Nuclear-Encoded tRNAs and Proteins Do Not Use the Same Import Pathway to Enter the Mitochondria.
To investigate the relationship between protein import and tRNA import, we studied in parallel the effect of the mitochondrial presequence of ATP synthase F1β subunit from Nicotiana plumbaginifolia on the import of GluRS-GFP and tRNAAla transcript. The F1β presequence was previously shown to efficiently inhibit the import of various precursor proteins into isolated mitochondria (21). Here, preincubation of mitochondria with 3 μM F1β presequence efficiently inhibited GluRS-GFP import, with complete inhibition at 5 μM (Fig. 4A). By contrast, addition of similar amounts of F1β presequence has no effect on the import of tRNAAla transcript into mitochondria (Fig. 4B). Second, unlabeled tRNAAla transcript was used as a competitor in tRNA import or protein import assays performed in parallel. Whereas addition of increasing concentrations of unlabeled tRNAAla transcript progressively blocks by competition the internalization of the homologous-labeled tRNAAla transcript (Fig. 4C), no inhibition of GluRS-GFP occurs when increasing amounts of unlabeled tRNAAla transcript were added to the assay (Fig. 4D). In summary, these results show that tRNAs and proteins are imported into mitochondria via distinct pathways.
Discussion
This article presents evidence that the VDAC located on the outer membrane of plant mitochondria is important for translocation of cytosolic tRNAs into plant mitochondria. It also provides evidence that components of the TOM complex play an essential role in the tRNA import process. However, even if the tRNA and protein import machineries share common components, our data demonstrate that tRNAs and proteins are translocated via distinct pathways in plant mitochondria.
The passage of cytosolic tRNAs across the mitochondrial outer membrane is trypsin sensitive and is likely initiated by import receptors at the surface of the organelle. Antibodies against TOM20 and TOM40, two protease-sensitive outer membrane proteins of the TOM complex, inhibit tRNA binding onto mitochondria. This finding supports the involvement of at least part of the TOM complex in the initial fixation of tRNAs at the surface of mitochondria. A subsequent question is whether tRNAs directly interact with these proteins. Indeed, the requirement of other factors (belonging or not to the TOM complex) cannot be excluded and their possible identification awaits further work. In yeast, there are several lines of evidence showing that TOM20 interacts with the positively charged and hydrophobic regions of mitochondrial presequences (22). By contrast, tRNAs represent negatively charged macromolecules and, up to now, interactions between nucleic acids and TOM proteins have never been detected to our knowledge. It is, however, worth noting that plant TOM complex differs significantly from the yeast and mammalian homologues (23). In particular, in plants, although it has been recently shown that there is a convergent evolution of the distinct plant and animal TOM20 receptors to a common function (protein import into mitochondria) (24), plant TOM20 receptors differ considerably in sequence from the fungal and animal TOM20 homologs. In fact, the yeast TOM complex has a negative net charge, whereas the Arabidopsis one has a positive net charge (25). It has been postulated that these differences in plant TOM complex are caused by the presence of a second protein translocation apparatus in plastids that allows better differentiation between these two systems. We can also hypothesize that in plants the TOM complex has evolved in relation to its function in tRNA import. Therefore, an important subject to be addressed in future studies will be understanding the basis of the interaction of tRNA with the TOM complex components. As mentioned above, tRNA import is an ATP-dependent process. Now, we also show that ATP is required for the tRNA binding step at the surface of mitochondria (Fig. 3B). The requirement of external ATP has been shown in all systems studied to date but its precise role at the outer membrane is still unknown. One can hypothesize that recognition by import receptors and translocation through the outer membrane channel require at least a partial unfolding of the secondary or tertiary structure of the tRNA by helicases and/or chaperones that are known to require ATP hydrolysis for their activity.
After tRNA binding at the mitochondrial surface by import receptors occurs a translocation step through VDAC. This porin is the major component of the mitochondrial outer membrane and forms a voltage-gated pathway by which negatively charged metabolites such as succinate, malate, or ATP cross the membrane. In addition, VDAC has been implicated in other important functions such as compartmentation or apoptosis (26). Because the molecular cutoff of the VDAC is ≈3 kDa, it was commonly believed that VDAC is only permeable to small molecules and ions. However, during the last decade several lines of evidence raised questions about the pore size of the VDAC protein in physiological membranes and the control of its permeability (27). Most interestingly, DNA was successfully translocated across a planar bilayer membrane containing isolated mammalian mitochondrial VDAC (28). Moreover, it has been shown that plant mitochondria can import DNA via VDAC (18), and very recently it was reported that DNA can be imported into human and rat mitochondria (29). Our data now provide evidence that VDAC can fulfill another essential function: the transport of tRNAs into plant mitochondria. Indeed, with a 2- to 4-nm pore in its open state, VDAC can permit the passage of nucleic acids, and this diameter is compatible with the double-helix minimal diameter of DNA (30, 31). However, because of their L-shaped structure, tRNAs appear to be larger and the question of whether they must be unfolded to allow translocation through VDAC remains to be answered. In the future identification of the domain(s) of plant VDAC interacting with tRNAs and characterization of its crystallographic structure will help to understand how tRNAs can be translocated through its pore.
Finally, in plants, in vivo tRNA import is highly specific, and the number and identity of imported tRNAs vary from one plant to the other (32). The level of in vivo selectivity is such that cytosol-specific tRNAs are not significantly detectable in purified mitochondria (e.g. ref. 33). By contrast, the selectivity obtained during in vitro tRNA import experiments was either limited (14) or absent (34). Altogether, these data suggest that some specificity factors have been lost in the in vitro import assays. For instance, aminoacyl-tRNA synthetases, although likely implicated in the import process (35), are not present in our in vitro system, which might partially explain the reduced specificity. We also have to explain why a cytosolic tRNA, with a priori the same sequence, is imported in one plant but not in the other (34). This tricky question awaits future research. Concerning VDAC, similar interactions were obtained when Northwestern experiments were performed with either plant cytosolic-specific tRNAs or unrelated transcripts. Competition experiments also showed that poly(U), oligonucleotide, or linearized pBluescript vector are able to block by competition interaction between labeled tRNAAla transcript and VDAC as efficiently as unlabeled tRNAAla transcript (data not shown). These observations are in agreement with the great variety of substrates (e.g., various metabolites, nucleotides, DNA) already shown to be transported via VDAC. It thus seems likely that VDAC, by itself, is not responsible for tRNA import selectivity and other factors and/or regulatory steps must be involved.
So far, based on data obtained with yeast and protozoans, two tRNA import models emerge. In the first model, soluble protein import factors, like the glycolytic enzyme enolase and aminoacyl-tRNA synthetases, and the protein import machinery are required for the import of tRNALys(CUU) into S. cerevisiae mitochondria (3). In the second model, a RNA-specific translocation apparatus is implicated without need of cytosolic factors. Indeed, in Leishmania, a still-unidentified, 15-kDa tRNA binding protein is apparently required for transfer across the outer membrane (36). Whether this putative receptor belongs to the TOM complex or not is an interesting question to answer in the future. Furthermore, an RNA import complex, including two bifunctional respiratory proteins, isolated from L. tropica inner mitochondrial membrane is implicated in tRNA import (12, 13). However, because of the conflicting data reported so far (see above), identification of import factors in other trypanosomatids might help in understanding the complexity of mitochondrial tRNA import. In view of our observations, nuclear-encoded tRNAs, for their transport into plant mitochondria, are first recognized by protein components of the TOM complex and then are translocated through a tRNA conducting channel including VDAC. However, whether tRNA transfer from the TOM complex to VDAC is direct or whether it requires intermediate steps with other protein components remains an open question. In our model, the general insertion pore that translocates proteins through the outer membrane is not involved in the tRNA translocation step. In the yeast model, tRNALys(CUU) is cotranslocated through the protein import machinery with a carrier, a precursor form of the mitochondrial LysRS (3). In that case, the role of the TOM complex in tRNALys import is indirect. In plants, external addition of cytosolic factors is not required for mitochondrial import of tRNAs in vitro and no obvious carrier is present in the medium. Therefore, the function of TOM proteins in tRNA import seems to be different between yeast and plants. Finally, the involvement of the glycolytic enzyme enolase in yeast, of respiratory proteins in Leishmania, and of the porin VDAC in plants shows that eukaryotic cells have likely developed independent mitochondrial tRNA import systems by recruiting various preexisting functional proteins. Whether these systems share components (for instance, the TOM complex) and whether other multifunctional proteins have to be recruited are questions that need further investigation.
Materials and Methods
Isolation of Plant Mitochondria, Preparation of Outer Mitochondrial Membranes, and VDAC Identification.
Mitochondria were isolated from potato tubers (16). Outer membranes were purified according to a modified version of a known protocol (15). Mitochondria (50 mg of mitochondrial proteins) were resuspended in 10 ml of swelling buffer (5 mM potassium phosphate, pH 7.2) and kept on ice for 10 min. The same volume of swelling buffer was added, and mitochondria were ruptured in a potter homogenizer. Outer membranes were separated from mitoplasts by centrifugation through a sucrose step gradient (60%–32%–15% sucrose in 1 mM EDTA, 1 mM PMSF, and 10 mM potassium phosphate, pH 7.2) at 4°C for 10 min at 125,000 × g. Outer membranes were collected from the 15%/30% interphase and diluted five times in washing buffer (0.3 M mannitol/1 mM EDTA/0.1% BSA/10 mM potassium phosphate, pH 7.2). Outer membranes were pelleted for 10 min at 170,000 × g. VDAC was purified from outer membrane proteins separated by SDS/PAGE, blotted onto Immobilon-P membrane (Millipore, Bedford, MA), and its N-terminal sequence was determined (473A sequencer; Applied Biosystems, Foster City, CA).
PCR Amplification, Cloning, and E. coli Overexpression of VDAC and GFP.
The cDNA encoding potato VDAC (15) and the GFP sequence (16) were amplified by RT-PCR, cloned into pQE60 vector, and used to overproduce and purify His-tag proteins according to the manufacturer's recommendation (Qiagen, Valencia, CA). For gel-shift assays, a refolding step was added on the Ni-NTA columns. It consisted of a three-step urea gradient (8, 4, and 0 M) in washing solution (5 mM MgCl2, 50 mM Tris·Maleate, pH 8.5) containing 2% octyl β-d-glucopyranoside.
Northwestern and Gel-Shift Experiments.
Northwestern experiments were performed essentially as described (37). Gel-shift assays were conducted as described for the Promega (Madison, WI) gel-shift assay system. For competition assays, 2- to 4- fold excess of unlabeled tRNAAla was included in the reaction.
In Vitro tRNA Import into Isolated Mitochondria and tRNA Binding Experiments onto Mitochondria.
Mitochondrial import of plant cytosolic tRNAAla transcript was carried out as described (14), except for the addition of 0.2 μl of RNase inhibitor (RNase Out; Invitrogen, Carlsbad, CA) in the import medium. To analyze the effect of antibodies, RuR, ATR, unlabeled tRNAAla transcript or F1β presequence, mitochondria were preincubated on ice for 10 min with the appropriate product before the addition of 32P-labeled in vitro tRNAAla transcript. Binding assays were performed under the same conditions as for tRNA import except that the RNase digestion step was omitted. Under these conditions, the ratio of imported tRNA/bound tRNA was estimated to be ≈0.1. All polyclonal antibodies tested on tRNA import were first checked for the presence of RNases in their antisera. To do so, upon incubation of labeled tRNAAla transcript in import medium for 20 min at 25°C and in the presence of antisera, RNA was extracted and analyzed on 15% denaturing polyacrylamide gel. For most antisera, 10–15% of degradation, relative to a mock treatment without antibodies, was calculated.
In Vitro Protein Import into Isolated Mitochondria.
Import of 35S-labeled fusion protein GluRS-GFP into purified potato mitochondria was performed as described (16). F1β presequence was isolated as described (21). To study the effect of antibodies, RuR, ATR, and F1β presequence, mitochondria were preincubated for 10 min on ice with the appropriate product before the addition of labeled GluRS-GFP.
Western Blot Analysis and Antibodies.
Western blot analyses were conducted according to standard procedures. Antibodies against potato mitochondrial TOM20 and VDAC, Vicia faba mitochondrial TOM40, wheat mitochondrial NAD9, and bean cytosolic LeuRS were kindly provided by H. P. Braun (Hannover University, Hannover, Germany), M. A. Harmey (University of Dublin, Dublin, Ireland), J. M. Grienenberger (Institut de Biologie Moléculaire des Plantes, Strasbourg, France), and A. Dietrich (Institut de Biologie Moléculaire des Plantes, Strasbourg, France), respectively.
Supplementary Material
Acknowledgments
We thank A. Cosset for technical assistance. This work was supported by the Centre National de la Recherche Scientifique, the Swedish Research Council (E.G.), and a fellowship from the French Ministère de l'Enseignement Supérieur et de la Recherche (to T.S. and L.D.).
Abbreviations
- ATR
atractyloside
- RuR
Ruthenium red
- TOM
translocase outer mitochondrial membrane
- VDAC
voltage-dependent anion channel.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Entelis NS, Kolesnikova OA, Martin RP, Tarassov IA. Adv Drug Deliv Rev. 2001;49:199–215. doi: 10.1016/s0169-409x(01)00135-1. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya SN, Adhya S. RNA Biol. 2004;1:84–88. doi: 10.4161/rna.1.2.1180. [DOI] [PubMed] [Google Scholar]
- 3.Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I. Genes Dev. 2006;20:1609–1620. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinehart J, Krett B, Rubio MA, Alfonzo JD, Söll D. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee S, Bhattacharyya SN, Adhya S. J Biol Chem. 1999;274:31249–31255. doi: 10.1074/jbc.274.44.31249. [DOI] [PubMed] [Google Scholar]
- 6.Yermovsky-Kammerer AE, Hajduk SL. Mol Cell Biol. 1999;19:6253–6259. doi: 10.1128/mcb.19.9.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nabholz CE, Horn EK, Schneider A. Mol Biol Cell. 1999;10:2547–2557. doi: 10.1091/mbc.10.8.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubio MA, Liu X, Yuzawa H, Alfonzo JD, Simpson L. RNA. 2000;6:988–1003. doi: 10.1017/s1355838200991519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider A, Maréchal-Drouard L. Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 10.Adhya S, Ghosh T, Das A, Kanti Bera S, Mahapatra S. J Biol Chem. 1997;272:21396–21402. doi: 10.1074/jbc.272.34.21396. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya SN, Chatterjee S, Goswami S, Tripathi G, Dey SN, Adhya S. Mol Cell Biol. 2003;23:5217–5224. doi: 10.1128/MCB.23.15.5217-5224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goswami S, Dhar G, Mukherjee S, Mahata B, Chatterjee S, Home P, Adhya S. Proc Natl Acad Sci USA. 2006;103:8354–8359. doi: 10.1073/pnas.0510869103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee S, Home P, Mukherjee S, Mahata B, Goswami S, Dhar G, Adhya S. J Biol Chem. 2006;281:25270–25277. doi: 10.1074/jbc.M604126200. [DOI] [PubMed] [Google Scholar]
- 14.Delage L, Dietrich A, Cosset A, Maréchal-Drouard L. Mol Cell Biol. 2003;23:4000–4012. doi: 10.1128/MCB.23.11.4000-4012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heins L, Mentzel H, Schmid A, Benz R, Schmitz UK. J Biol Chem. 1994;269:26402–26410. [PubMed] [Google Scholar]
- 16.Duchêne A, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters N, Zaepfel M, Maréchal-Drouard L, Small ID. Proc Natl Acad Sci USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gincel D, Zaid H, Shoshan-Barmatz V. Biochem J. 2001;358:147–155. doi: 10.1042/0264-6021:3580147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koulintchenko M, Konstantinov Y, Dietrich A. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki H, Kadowaki T, Maeda M, Sasaki H, Nabekura J, Sakaguchi M, Mihara K. J Biol Chem. 2004;279:50619–50629. doi: 10.1074/jbc.M408604200. [DOI] [PubMed] [Google Scholar]
- 20.Jänsch L, Kruft V, Schmitz UK, Braun HP. J Biol Chem. 1998;273:17251–17257. doi: 10.1074/jbc.273.27.17251. [DOI] [PubMed] [Google Scholar]
- 21.Moberg P, Nilsson S, Stahl A, Eriksson AC, Glaser E, Maler L. J Mol Biol. 2004;336:1129–1140. doi: 10.1016/j.jmb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Ryan MT, Wagner R, Pfanner N. Int J Biochem Cell Biol. 2000;32:13–21. doi: 10.1016/s1357-2725(99)00114-4. [DOI] [PubMed] [Google Scholar]
- 23.Whelan J, Schleiff E. In: Plant Mitochondria: From Genome to Function. Day DA, Millar H, Whelan J, editors. Boston: Kluwer; 2004. pp. 31–54. [Google Scholar]
- 24.Perry AJ, Hulett JM, Likic VA, Lithgow T, Gooley PR. Curr Biol. 2006;16:221–229. doi: 10.1016/j.cub.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Werhahn W, Jänsch L, Braun H-P. Plant Phys Biochem. 2003;41:407–416. [Google Scholar]
- 26.Al Bitar F, Roosens N, Smeyers M, Vauterin M, Van Boxtel J, Jacobs M, Homble F. Biochim Biophys Acta. 2003;1625:43–51. doi: 10.1016/s0167-4781(02)00590-0. [DOI] [PubMed] [Google Scholar]
- 27.Bathori G, Csordas G, Garcia-Perez C, Davies E, Hajnoczky G. J Biol Chem. 2006;281:17347–17358. doi: 10.1074/jbc.M600906200. [DOI] [PubMed] [Google Scholar]
- 28.Szabo I, Bathori G, Tombola F, Coppola A, Schmehl I, Brini M, Ghazi A, De Pinto V, Zoratti M. FASEB J. 1998;12:495–502. doi: 10.1096/fasebj.12.6.495. [DOI] [PubMed] [Google Scholar]
- 29.Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Hum Mol Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 30.Mannella CA, Forte M, Colombini M. J Bionenerg Biomembr. 1992;24:7–19. doi: 10.1007/BF00769525. [DOI] [PubMed] [Google Scholar]
- 31.Szabo I, Bathori G, Tombola F, Coppola A, Schmehl I, Brini M, Ghazi A, De Pinto V, Zoratti M. FASEB J. 1998;12:495–502. doi: 10.1096/fasebj.12.6.495. [DOI] [PubMed] [Google Scholar]
- 32.Duchêne A-M, Maréchal-Drouard L. Biochem Biophys Res Commun. 2001;285:1213–1216. doi: 10.1006/bbrc.2001.5303. [DOI] [PubMed] [Google Scholar]
- 33.Kumar R, Maréchal-Drouard L, Akama K, Small I. Mol Gen Genet. 1996;252:404–411. doi: 10.1007/BF02173005. [DOI] [PubMed] [Google Scholar]
- 34.Salinas T, Schaeffer C, Maréchal-Drouard L, Duchêne AM. Biochimie. 2005;87:863–872. doi: 10.1016/j.biochi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Delage L, Duchêne AM, Zaepfel M, Maréchal-Drouard L. Plant J. 2003;34:623–633. doi: 10.1046/j.1365-313x.2003.01752.x. [DOI] [PubMed] [Google Scholar]
- 36.Mahapatra S, Adhya S. J Biol Chem. 1996;271:20432–20437. doi: 10.1074/jbc.271.34.20432. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh A, Ghosh T, Ghosh S, Das S, Adhya S. Nucleic Acids Res. 1994;22:1663–1669. doi: 10.1093/nar/22.9.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.