Abstract
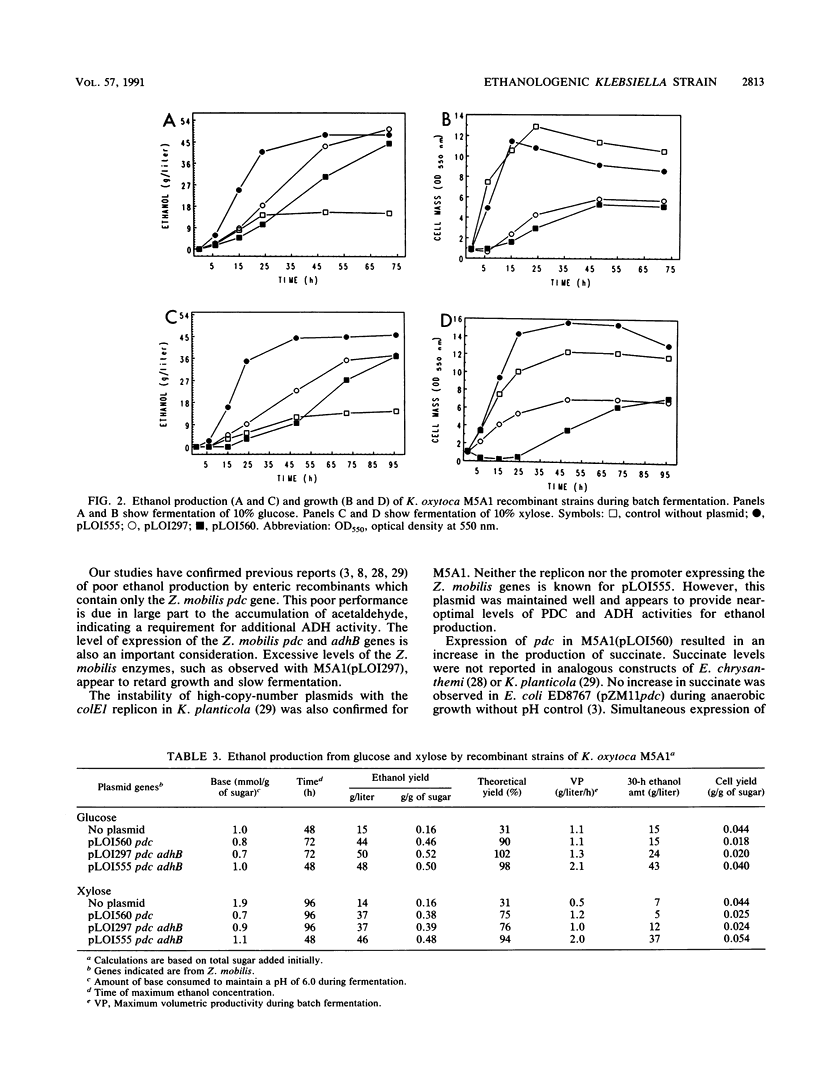
The efficient diversion of pyruvate from normal fermentative pathways to ethanol production in Klebsiella oxytoca M5A1 requires the expression of Zymomonas mobilis genes encoding both pyruvate decarboxylase and alcohol dehydrogenase. Final ethanol concentrations obtained with the best recombinant, strain M5A1 (pLOI555), were in excess of 40 g/liter with an efficiency of 0.48 g of ethanol (xylose) and 0.50 g of ethanol (glucose) per g of sugar, as compared with a theoretical maximum of 0.51 g of ethanol per g of sugar. The maximal volumetric productivity per hour for both sugars was 2.0 g/liter. This volumetric productivity with xylose is almost twice that previously obtained with ethanologenic Escherichia coli. Succinate was also produced as a minor product during fermentation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterthum F., Ingram L. O. Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl Environ Microbiol. 1989 Aug;55(8):1943–1948. doi: 10.1128/aem.55.8.1943-1948.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chen L. M., Maloy S. Regulation of proline utilization in enteric bacteria: cloning and characterization of the Klebsiella put control region. J Bacteriol. 1991 Jan;173(2):783–790. doi: 10.1128/jb.173.2.783-790.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989 Sep;5(3):223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A., Malpartida F., Kieser H. M., Ikeda H., Duncan J., Fujii I., Rudd B. A., Floss H. G., Omura S. Production of 'hybrid' antibiotics by genetic engineering. Nature. 1985 Apr 18;314(6012):642–644. doi: 10.1038/314642a0. [DOI] [PubMed] [Google Scholar]
- Ingram L. O., Conway T., Clark D. P., Sewell G. W., Preston J. F. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987 Oct;53(10):2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L. O., Conway T. Expression of Different Levels of Ethanologenic Enzymes from Zymomonas mobilis in Recombinant Strains of Escherichia coli. Appl Environ Microbiol. 1988 Feb;54(2):397–404. doi: 10.1128/aem.54.2.397-404.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAHL M. C., WILSON P. W., FIFE M. A., EWING W. H. NITROGEN FIXATION BY MEMBERS OF THE TRIBE KLEBSIELLEAE. J Bacteriol. 1965 Jun;89:1482–1487. doi: 10.1128/jb.89.6.1482-1487.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N., Nakagawa M., Kobayashi K., Yamano S., Izawa Y., Nakamura K., Harashima K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990 Dec;172(12):6704–6712. doi: 10.1128/jb.172.12.6704-6712.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Alterthum F., Ingram L. O. Effects of environmental conditions on xylose fermentation by recombinant Escherichia coli. Appl Environ Microbiol. 1990 Feb;56(2):463–465. doi: 10.1128/aem.56.2.463-465.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Beall D. S., Mejia J. P., Shanmugam K. T., Ingram L. O. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991 Apr;57(4):893–900. doi: 10.1128/aem.57.4.893-900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka A., Rackwitz H. R., Frischauf A. M., Hohn B., Lehrach H. Selective isolation of cosmid clones by homologous recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4129–4133. doi: 10.1073/pnas.81.13.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S. C., Voige W. H., Dennis D. E. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-beta-hydroxybutyrate biosynthetic pathway. J Bacteriol. 1988 Oct;170(10):4431–4436. doi: 10.1128/jb.170.10.4431-4436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolan J. S., Finn R. K. Fermentation of d-Xylose and l-Arabinose to Ethanol by Erwinia chrysanthemi. Appl Environ Microbiol. 1987 Sep;53(9):2033–2038. doi: 10.1128/aem.53.9.2033-2038.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolan J. S., Finn R. K. Fermentation of d-Xylose to Ethanol by Genetically Modified Klebsiella planticola. Appl Environ Microbiol. 1987 Sep;53(9):2039–2044. doi: 10.1128/aem.53.9.2039-2044.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]