Abstract
Hereditary tyrosinemia type I and alkaptonuria are disorders of tyrosine catabolism caused by deficiency of fumarylacetoacetate hydrolase (FAH) and homogentisic acid dioxygenase (HGD), respectively. Tyrosinemia is a severe childhood disease that affects the liver and kidneys, but alkaptonuria is a more benign adult disorder in comparison. Because HGD is upstream of FAH in the tyrosine pathway, mice doubly mutant in both enzymes were found to be protected from the liver and renal damage of tyrosinemia as hypothesized. Mice mutant at the tyrosinemic locus but heterozygous for alkaptonuria spontaneously developed clonal nodules of functionally normal hepatocytes that were able to rescue the livers of some mice with this genotype. This phenotypic rescue was a result of an inactivating mutation of the wild-type homogentisic acid dioxygenase gene, thus presenting an example of an in vivo suppressor mutation in a mammalian model.
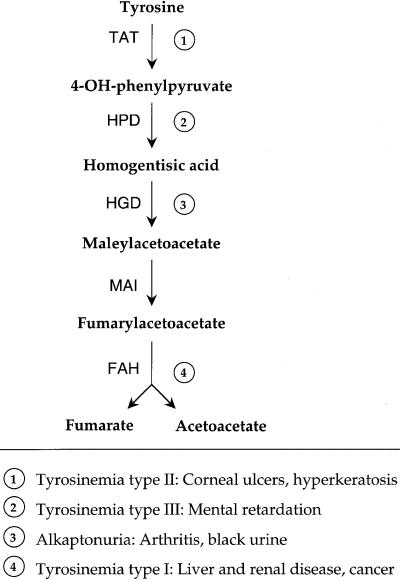
Hereditary tyrosinemia type I (HT1) is an inborn error of metabolism caused by deficiency of fumarylacetoacetate hydrolase (FAH), the enzyme that carries out the last step of the tyrosine catabolic pathway (Fig. 1) (1, 2). Of the four genetic diseases known to result from deficiencies in specific enzymes in the pathway, HT1 is the most severe and is characterized by progressive liver dysfunction and renal tubular disease (2). Moreover, HT1 is unique among the tyrosine catabolic pathway diseases in that patients have a high risk for developing liver cancer (3). Maleylacetoacetate (MAA) and fumarylacetoacetate (FAA), the two compounds that precede FAH in the pathway and, thus, accumulate in FAH-deficient hepatocytes are predicted to be alkylating agents by structure and are the primary cause of the liver damage (1, 4). Currently, the drug NTBC (2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione), which blocks the pathway upstream of the formation of either MAA or FAA by inhibiting 4-hydroxyphenylpyruvate dioxygenase (HPD) (Fig. 1), is used to treat patients with HT1 (5). However, NTBC has the disadvantage of markedly elevating blood tyrosine levels and, therefore, needs to be combined with dietary restriction of phenylalanine and tyrosine, because increased tyrosine may lead to corneal ulcers and neurological problems as is seen in patients with hereditary tyrosinemia types II and III (Fig. 1) (2). Additionally, NTBC may not prevent hepatocarcinomas in patients with HT1 (3, 6).
Figure 1.
Tyrosine-degradation pathway and its diseases.
Deficiency of another enzyme upstream of FAH in the pathway, homogentisate 1,2 dioxygenase (HGD), causes alkaptonuria (aku), which has no increase in tyrosine levels (7). Instead, there is an accumulation of the substrate for HGD, homogentisic acid, which is excreted in the urine and can be oxidized spontaneously to a black pigment, causing darkly colored urine (8). This pigment also is deposited in many tissues, causing ochronosis, arthritis, and connective tissue damage in adulthood (7).
Because elevated blood tyrosine and liver cancer are not found in aku patients, and the toxic compounds MAA and FAA do not accumulate, we hypothesized that inhibition of the pathway at HGD may be a better treatment for HT1 patients than blocking at HPD by using NTBC. This hypothesis could be tested by using the mouse models of aku and HT1. An aku mouse model has been described that was created by N-ethyl-N-nitrosourea mutagenesis (9). Aku mice were found to have a splice mutation, causing the production of two shorter transcripts that predict severely truncated HGD proteins (10). These mice have high levels of homogentisic acid in the urine; however, older mice show no evidence of ochronosis or joint disease, possibly because mice can synthesize ascorbic acid, unlike humans (9). A knock-out mouse model of HT1 also exists (FAHΔexon5) (11), which is lethal unless kept on NTBC (12). When taken off NTBC, FAHΔexon5 mice experience liver failure, showing necrosis, inflammation, and dysplastic cells histologically. They also exhibit renal tubular disease and will develop hepatic cancer even while on NTBC (12). By breeding these two mouse models, we tested the hypothesis that a block in the pathway at HGD could rescue FAH knock-out mice from the severe HT1 phenotype.
Materials and Methods
Strains of Mice and Animal Husbandry.
Aku mice were obtained from Xavier Montagutelli at the Pasteur Institute (Paris). Aku and FahΔexon5 mice (11) were bred to create double heterozygotes, which then were crossed to obtain the desired genotypes (without controlling for strain background). Pups were genotyped at the Fah locus by using a 3-primer PCR on 200 ng of tail-cut DNA as described (11). Aku mice were genotyped as described by PCR (10). All breeders and all mutant animals were treated with NTBC containing drinking water at a concentration of 7.6 mg/liter (provided by S. Lindstedt, Gottenburg, Sweden). FAH mutants with varying Hgd genotypes were kept on NTBC until an age of at least 2 months, when NTBC was removed. Weights were followed at a weekly basis, and animals were sacrificed at 4 weeks after stopping NTBC or at a later time point for those that survived NTBC withdrawal for 7 or more weeks. Animal care and experiments were all in accordance with the Guidelines of the Department of Animal Care at Oregon Health Sciences University.
HGD Enzyme Assay.
Liver was homogenized and then sonicated for 2 × 10 sec in 500 μl of 0.25 M sucrose. The liver extract then was clarified by centrifugation (full speed in a microcentrifuge for 15 min at 4°C), and 50–200 μg of protein was added to the reaction mixture containing 100 mM potassium phosphate buffer, pH 7/2 mM ascorbate/50 μM iron sulfate/250 μM homogentisate. The production of maleylacetoacetate was measured spectrophotometrically at 330 nm, as described (13, 14).
Biochemical Analysis and Quantitative Amino Acid Analysis.
Individual plasma samples of 25 μl were analyzed on a Beckman 6300 automated amino acid analyzer (15). Samples from animals were obtained as follows. Animals were killed by decapitation and blood was collected. For anticoagulation, the blood was mixed immediately with 10 μl of sodium-heparin. The red blood cells were removed by a brief centrifugation, and the plasma was frozen at −80°C. Twenty microliters of plasma were mixed with 80 μl of a solution of 7% BSA and assayed for alanine aminotransferase, bilirubin, and creatinine levels with a Kodak Ektachem 700 chemistry analyzer.
Histology and Immune Histology.
For immunohistology, tissues were fixed in 10% phosphate-buffered formalin, pH 7.4, dehydrated in 100% ethanol, and embedded in paraffin wax at 58°C. Five-micron sections were rehydrated and stained with hematoxylin/eosin (H&E). Endogenous peroxidase activity was blocked with 3% H2O2 and methanol. Avidin and biotin pretreatment was used to prevent endogenous staining. Rabbit antibody to mouse AFP was obtained from ICN (cat no. 64-561) and used at a dilution of 1:10,000. Rabbit antibody to mouse ki-67 was obtained from Novocastra (NCL-ki67p) and used at a dilution of 1:300; the antigen–antibody complex was detected with a goat anti-rabbit antibody from Chemicon (AQ 132B), which was used at a dilution of 1:100. Color development was performed with the AEC detection kit from Ventana Medical Systems (Tucson, AZ).
Reverse Transcription–PCR (RT-PCR).
At the time of sacrifice, livers from Fah−/−, Hgdaku/Hgdwt mice were dissected into random pieces of ≤5 mm in diameter. In one animal sacrificed at 7 weeks off NTBC, individual nodules 1–2 mm in size were identifiable, and 10 of these were isolated carefully. Each piece was homogenized directly in RNAzol B (Tel-Test, Friendswood, TX), and total cellular RNA was isolated (16). Ten micrograms of total cellular RNA was reverse-transcribed with 0.8 μg oligo(dT) in a volume of 40 μl by using a BRL kit (Life Technologies, Gaithersburg, MD). Two microliters of the RT reaction was subjected to PCR amplification under the following conditions: 94°C for 5 min, followed by 30 cycles of 90°C × 30 sec, 55°C × 30 sec, and 72°C × 1 min, and then a final extension step of 72°C × 10 min. GeneAmp 10× PCR buffer and 2.5 mM Mg2+ (Perkin–Elmer) were used in a 25-μl reaction with either Taq DNA Polymerase (Boehringer Mannheim) or Pfu DNA Polymerase (Stratagene). Primer positions are shown in Fig. 4; the reverse primer at position 1535 in the 3′ UTR of Hgd (17) had the sequence 5′-TCAATTACAGTAGAGGGCTCCAGTC-3′, and the forward primer located at position 866 had the sequence 5′-TAATACGACTCACTATAGGGGCCTGGTATGAAGATCG-3′.
Figure 4.
Schematic representation of murine Hgd. Wild-type and aku alleles are shown. M1, splice mutant allele with one exon deleted. M2, splice mutant allele with two exons deleted. Positions of RT-PCR primers used for mutation analysis of Hgd also are shown.
Cloning and Sequencing.
The PCR products derived from the 3′ half of Hgd were separated by electrophoresis to isolate the wild-type-length product (see Fig. 4). This fragment was isolated by using a QIAquick gel extraction kit (Qiagen) and then cloned with either AdvanTAge PCR cloning kit (CLONTECH) or a PCR-Script kit for Pfu products (Stratagene). PCR was performed on the resulting bacterial colonies by using the same PCR protocol as described above, except only 25 cycles were used. Dideoxy sequencing (18) by using the same forward primer as listed above then was performed on the PCR products after cleaning with a QIAquick PCR purification kit (Qiagen).
Allele-Specific Oligonucleotide (ASO) Hybridization.
Fourteen- to 15-bp oligonucleotides homologous to the wild-type and various mutant sequences were designed (19). Ten microliters of the PCR product from PCR-2 clones (from colonies, as described above) was denatured in 100 μl 0.4 M NaOH/25 mM EDTA and spotted onto a Hybond-N+ membrane (Amersham) in a Schleicher & Schuell dot-blot apparatus. Five picomoles of each ASO was end-labeled by T4 polynucleotide kinase and [γ-32P]ATP. The kinase was heat-inactivated, and 50 pmol of the opposing, unlabeled ASO was added. The dot blots were hybridized overnight at 25–45°C (depending on the calculated Tm) in 6× SSPE/1% SDS/5× Denhardt’s solution. Washes were carried out for 10–30 min with either 5× or 2× SSC and 0.5% SDS at 25–42°C.
Restriction Enzyme Mutation Analysis.
Three mutations were confirmed by altering a base pair in a PCR primer near the mutation to create a restriction site. For mutation A → G at 998, primer 5′-ATGGAAACTACACACCCTTC-3′ created a TaqI site recognizing the mutant sequence. For mutation A → C at 1154, primer 5′-GTAATAAGGAGGTCTGAACG-3′ created a Psp 14061 (Boehringer Mannheim) site for the wild-type sequence. Lastly, for mutation C → T at 1155, primer 5′-GTGGGGAGTTGCAGATAAGA-3′ created a BglII site for mutant clones only.
Quantitative PCR Genotyping.
To determine the ratio of wild-type and mutant alleles in genomic DNA, 70 ng of DNA was isolated from separate, 5-mm cubes of Fah−/−, Hgdaku/Hgdwt mouse livers as described (20) and used for PCR. Two hundred nanograms of the forward primer was end-labeled by T4 polynucleotide kinase and [γ-32P]ATP and then centrifuged through a microcon-3 (Amicon) to remove unincorporated nucleotides. Approximately 10 ng of the labeled primer was added to each PCR. PCR and digestion with RsaI were performed as described previously (10). Products were run on a 2.5% agarose gel, which then was dried and placed on a phosphorimaging screen for 2 hr. Results were analyzed by using iplab gel software (Scanalytics, Fairfax, VA).
Because only two-thirds of the liver genomic DNA consists of hepatocyte DNA (21), and because the phenotypic reversion occurred only in these cells, the true ratio of mutant/wild-type DNA was calculated by using the following formulas: X = 1/3 + 2/3 (m/w); m + w = 1, where X = measured ratio of mutant (m) over wild-type (w) DNA and m/w = ratio of mutant/wild-type DNA in hepatocytes only. The corrected ratios are given in Fig. 5B.
Figure 5.
Loss of heterozygosity by gene conversion or gene deletion. (A) RT-PCR on clonal nodules in a Fah−/−, Hgdaku/Hgdwt mouse. Only 4 of the 10 nodules showed a wild-type-length PCR product (∗). Mutant PCR products with one or two exons deleted (see Fig. 4) were amplified in each nodule. In those nodules that had an amplifiable wild-type allele, sequencing of cloned RT-PCR products showed the same mutation in all clones, confirming that the dissected sections indeed were clonal nodules. (B) PhosphorImager analysis of genotyping assay on liver genomic DNA. Two liver pieces (nonclonal) from three Hgd heterozygous mice were analyzed, as well as two heterozygous controls in which the wild-type allele represents approximately 50% of the signal, as expected. The percentage of wild-type allele calculated from the data is corrected to include hepatocyte DNA only (see Materials and Methods).
Results
Cross-Breeding of aku and HT1 Mice.
FAH knock-out mice were bred with aku mice to generate mice that are mutant at the Fah locus and wild-type, heterozygous, or mutant at the Hgd locus. These mice were kept on NTBC until age 2–4 months. NTBC then was discontinued and mice were sacrificed 4 weeks later. As predicted, Fah−/−, Hgdaku/Hgdaku (double-mutant) mice retained a healthy weight and appearance throughout the 4 weeks off NTBC, whereas Fah−/−, Hgdaku/Hgdwt (Hgd heterozygote) and Fah−/−, Hgdwt/Hgdwt (Hgd wild-type) mice lost weight and became sick (data not shown). At the time of sacrifice, the liver was pale and jaundiced in all but the double-mutant mice, in which the liver appearance was normal. Liver function tests, plasma amino acid levels, and liver histology were analyzed to confirm these macroscopic observations. Some double-mutant mice also were kept off NTBC for long-term study (12–14 months) of liver and kidney function to compare with those from only 4 weeks off NTBC.
Liver Function and Amino Acid Levels.
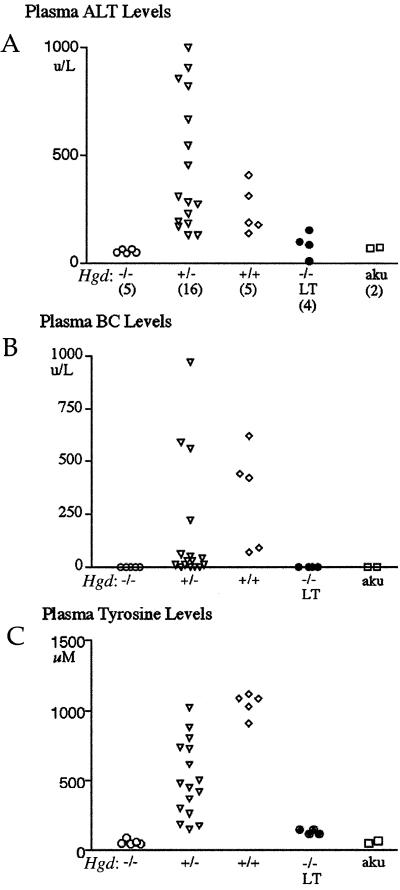
Results of liver function tests of double-mutant mice confirmed that plasma levels of alanine aminotransferase (Fig. 2A) and conjugated bilirubin (BC) (Fig. 2B) were normal in both short-term (4 weeks) and long-term periods off of NTBC. However, both liver function tests of Hgd heterozygous and Hgd wild-type mice were abnormal. Also, plasma tyrosine levels were in the normal range in double-mutant mice both long and short term, whereas tyrosine levels in Hgd heterozygous and Hgd wild-type mice were elevated because of the liver dysfunction in FAH deficiency (Fig. 2C) (12). Similar results were found for other plasma amino acid levels measured for each mouse genotype (data not shown). Aku mice that are wild type at the Fah locus also were included as normal controls. Thus, blockage of the tyrosine catabolic pathway at the level of HGD completely protected hepatic function in tyrosinemic mice, but did not increase blood tyrosine levels. Interestingly, liver function tests and amino acid levels were near normal in some of the Hgd heterozygous mice, indicating that they were not as severely affected by the liver disease as the Hgd wild-type mice.
Figure 2.
Liver-function tests and plasma amino acid levels for Fah−/− mice with varying Hgd genotypes. (A) ALT (alanine aminotransferase) levels. (B) BC (conjugated bilirubin) levels. (C) Plasma tyrosine levels. Double-mutant mice (−/−) are shown with both short-term (4-week) and long-term (LT) (12- to 14-month) periods off of NTBC. +/−, Hgd heterozygote; +/+, Hgd wild type; aku, alkaptonuria controls (Fah wild type). Numbers of mice from each genotype used throughout this study are given in parentheses.
Histology Results.
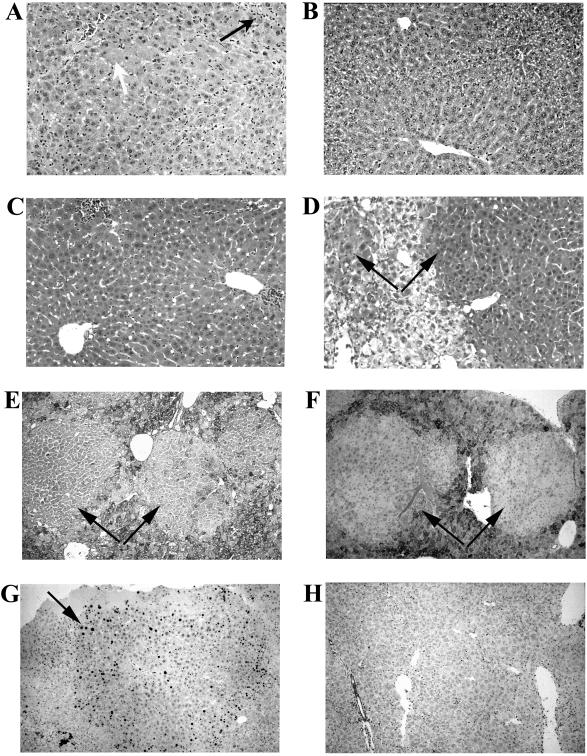
Fig. 3 A–D shows liver H&E staining of double-mutant, Hgd heterozygote, and Hgd wild-type mice. The latter two genotypes showed evidence of liver disease, with inflammation, necrosis, and dysplasia. The double mutants, however, had hepatocytes of homogeneous size with few inflammatory cells in both short- and long-term periods off of NTBC. Interestingly, all Hgd heterozygous mice had nodules of healthy-appearing hepatocytes that clearly were shown in the liver sections and often visible macroscopically at the time of sacrifice. The nodules appeared as distinct regions of normal hepatocytes that were low in glycogen and negative for ∝-fetoprotein, although both markers were elevated in the surrounding tyrosinemic hepatocytes. The nodules also stained positive for Ki67, a marker for proliferating cells (Fig. 3 E–G) (22). These findings confirmed that the nodules consisted of healthy, dividing hepatocytes. The presence of nodules in the livers of Hgd heterozygous mice explained why some of the liver-function tests and plasma amino acid levels were near normal in these mice, because the nodules created a population of functional hepatocytes in their liver.
Figure 3.
Liver histology. (A) H&E of Fah−/−, Hgdwt/Hgdwt mouse showing pathology typical of HT1, with many inflammatory cells (black arrow), necrotic cells (white arrow), and disorganized liver architecture, compared with (B) H&E for a double-mutant mouse 4 weeks off NTBC showing normal liver cells, with organized, columnar appearance of hepatocytes. (C) H&E of double-mutant mouse >1 year off NTBC also demonstrating normal liver histology. (D) H&E of Fah−/−, Hgdaku/Hgdwt, depicting nodules (black arrows) of healthy hepatocytes against a background of sick cells. Nodules were depleted of glycogen (PAS staining) (E), negative for ∝-fetoprotein (F), and contained many cells positive for Ki67 (G) whereas Hgd wild-type cells did not (H).
At the time of sacrifice, the kidneys in the double-mutant mice that were off NTBC for more than a year were of normal size and color. Histology results also showed that kidneys of the double-mutant mice were normal, with uniform proximal and distal tubules, whereas mice heterozygous or wild type at the Hgd locus showed the expected tubular dilatation and inflammation seen in an FAH knock-out mouse (12) (data not shown).
Origin of Nodules in HGD Heterozygotes.
Histology examination of all the mice sacrificed at 4 weeks off NTBC showed the presence of nodules in all of the Hgd heterozygous mice, but not in Hgd wild-type mice. Because the experiments described above had shown double-mutant hepatocytes to be metabolically normal, we hypothesized that loss of heterozygosity had occurred at the Hgd locus in these nodules by an inactivating mutation of the remaining wild-type Hgd allele. Because previous work has shown FAA to be mutagenic (4), mutations in Hgd could be induced by the accumulation of the alkylating substrate of FAH, FAA (and MAA), resulting in a double-mutant cell that is able to proliferate and form a healthy nodule in the liver. To determine whether mutation of the remaining wild-type Hgd allele could lead to a complete rescue of the FAH mutant phenotype, a cohort of mice that were Fah−/−, Hgdaku/Hgdwt were taken off NTBC. Five of 30 animals survived the NTBC withdrawal and recovered their normal weight. Three mice were sacrificed after being off NTBC for 7, 9, or 11 weeks. At the time this paper was submitted, the other two Hgd heterozygotes were still alive off of NTBC and appeared healthy.
At harvest, the livers of two Hgd heterozygous mice (from 9 and 11 weeks off NTBC) appeared normal, with no distinct nodules visible macroscopically or in histology sections. Liver-function tests also showed that the livers of these two mice were functioning normally (data not shown). Their kidneys, however, were pale and enlarged and showed extensive tubular damage histologically (data not shown). The liver of each animal was dissected into several pieces of about 5 mm in diameter, and RNA was isolated from each aliquot separately. RT-PCR then was performed on the resulting cDNA (PCR-1) by using primers to amplify the 3′ half of Hgd, where the wild-type allele can be separated from the splice variants (Fig. 4). The wild-type-length PCR product was isolated from an agarose gel and cloned. Five to 10 of these clones from each separate liver section were sequenced and putative mutations were identified. Liver sections from each of the three mice also were tested for HGD enzyme activity and had no HGD enzyme detectable (data not shown).
Mutation Analysis.
A second, independent RT-PCR (PCR-2) also was performed on each cDNA pool to differentiate real mutations from PCR errors. Any mutations identified in PCR-1 that would create obvious disruptions in the HGD protein, such as deletions and insertions, were confirmed in PCR-2 clones. Any sequence changes that caused a change in an amino acid were analyzed further only if they were in a conserved residue between Aspergillus nidulans and human HGD proteins (23). As indicated in Table 1, the Hgd mutations were confirmed by various methods from 90 clones sequenced in a total of 14 liver sections from the three mice. Eighteen sequence changes from PCR-1 were not present in the 50 independent clones from PCR-2 and then were assumed to be PCR errors or rare mutations that could not be confirmed.
Table 1.
Total confirmed Hgd mutations
| Deletions (9) | Insertions (3) | Missense (10) |
|---|---|---|
| ΔTCG at 966 (V) (ASO) | T at 1351 (ASO) | TGG → GAC (V → G, A → P) at 969 (RE) |
| ΔG at 1287 (ASO) | CG at 1363 (ASO) | T → G (W → G) at 974 (RE) |
| ΔG at 1292 (ASO) | 27-bp direct repeat (1261–1287) (seq.) | A → G (K → E) at 998 (RE*) |
| ΔC at 1330 (RE) | G → A (D → N) at 1052 (RE) | |
| ΔC at 1351 (RE) | A → C (T → P) at 1154 (RE*) | |
| ΔAT at 1367 (seq.) | C → T (T → I) at 1155 (RE*) | |
| ΔCTTC at 1380 (ASO) | A → G (R → G) at 1160 (RE) | |
| Δ31 bp (1253–1283) (seq.) | G → T (E → D) at 1195 (ASO) | |
| Δ27 bp (1458–1484) (seq.) | C → G (H → D) at 1283 (seq.) | |
| G → A (G → S) at 1286 (seq.) |
Methods of mutation confirmation are noted. Mutations were confirmed in PCR-2 clones by restriction digest (RE) if the mutation altered a restriction site. Otherwise, ASO hybridization was used when no restriction sites were altered by the mutation. Some mutations also were confirmed by creating a restriction site that recognized either the wild-type or mutant allele by altering a base in a PCR primer (RE*) (see Materials and Methods). Large alterations or mutations from clonal nodules (third mouse; see text) could be confirmed by sequencing alone (seq.). In the missense column, amino acid changes resulting from indicated base changes are referred to in parentheses.
Loss of Heterozygosity by Gene Conversion or Deletion.
The liver of the Hgd heterozygous mouse sacrificed at 7 weeks off NTBC showed some macroscopic nodularity and was dissected carefully in an attempt to isolate clonal nodules. Ten nodules were dissected, and RT-PCR was performed. Interestingly, the wild-type-length PCR products could be amplified only in 4 of these 10 nodules (Fig. 5A). We hypothesized that this lack of the wild-type-length product was due to gene conversion or deletion of the wild-type Hgd allele in these nodules. To confirm this hypothesis, PCR was performed on genomic DNA from two separate liver sections from each of the three mice. The genotyping assay developed for the aku mice (10) was used to separate the wild-type allele from the mutant Hgd allele by restriction digest with RsaI. One of the primers was labeled radioactively to allow quantitation of the results (Fig. 5B). Indeed, four of the six liver sections tested showed a reduced quantity of the wild-type band (<40%), as expected if gene conversion or deletion of the wild-type Hgd allele had occurred. A correction was applied to the ratios for all of the liver sections shown in Fig. 5B to include only hepatocyte DNA (see Materials and Methods), the only liver cells in which the entire tyrosine catabolic pathway is expressed (2). One liver section from mouse 2 had only 25% of the signal represented by the wild-type band (Fig. 5B), thus showing that gene conversion or deletion of the wild-type Hgd allele occurred in approximately 50% of the hepatocytes in this liver section.
Discussion
An engineered genetic block at HGD protected FAH-deficient mice from the severe liver disease, renal disease, and liver cancer found in the HT1 phenotype. This was shown with mice that were mutant at both Fah and Hgd loci, in both short-term analysis (4 weeks off NTBC) and in mice off NTBC for more than a year. In addition, double-mutant mice did not have the elevated tyrosine levels seen in mice treated with NTBC, the small molecule inhibitor of HPD (12). Therefore, we conclude that a pharmacologic block at HGD may be a better therapy for HT1 patients than NTBC, because no dietary restrictions for tyrosine and phenylalanine would be required, and liver cancer may be prevented. This approach also is not completely problem-free, because of the joint disease that can be caused by homogentisic acid accumulation. Unfortunately, a pharmacological inhibitor of HGD has not yet been reported.
Previously, others have rescued mice deficient in FAH by using a murine model for HPD deficiency (24). Endo et al. showed that mice mutant at both the Fah and Hpd loci had normal liver and kidney function, similar to our mice doubly mutant for FAH and HGD. However, the FAH/HPD-deficient mice had markedly elevated blood tyrosine, whereas the FAH/HGD-deficient mice did not. Also, in contrast to our report here, the formation of healthy nodules in the livers of Fah−/−, Hpd+/− mice was not reported. Hepatocellular carcinomas were not seen in either double-mutant mouse models, unlike FAH knock-out mice treated with NTBC, which do develop liver cancer. This result suggests that the lack of complete protection by NTBC in FAH knock-out mice must be due to an insufficient amount of drug to completely inhibit HPD in vivo.
Here, we have shown that a somatic mutation in a second gene (Hgd) in the tyrosine pathway can rescue the lethal liver disease of FAH knock-out mice. Such suppressor mutations previously have been reported in simple-model organisms such as yeast and Escherichia coli (25). In fact, a similar finding was discovered in the Aspergillus model of FAH deficiency, where suppressor mutations in Hgd also were found to prevent toxicity caused by growth of the FAH mutant strain on phenylalanine-containing medium (26). However, here we show an example of a suppressor mutation in vivo in a mammal.
Recently, an increasing number of examples of somatic reversion have been documented in regenerating tissues with genetic disease (27–31). The systems involved include the bone marrow, liver, and skin. In the cases of “classical” somatic reversion, one mutant allele is restored to wild-type function. The most common mechanism is intragenic mitotic recombination (29, 30). However, actual removal of point mutations and compensatory frameshifts also have been reported (27, 28, 32). We have reported previously that the selective growth advantage of genetically corrected cells can lead to liver repopulation with healthy hepatocytes and complete restoration of liver function (33). Here, we show that second-site mutations in other genes in the same pathway also can result in spontaneous liver repopulation and full phenotypic rescue. This mechanism may be more common than mutation reversion, especially in heterozygous carriers of the suppressor mutation, because any loss of function mutation will result in rescue, whereas the classic somatic reversion requires rare genetic events that restore gene function. In vivo suppressor mutations should be considered in cases of spontaneous phenotypic improvement of patients with genetic diseases involving the blood, liver, or skin.
Previous studies on the specific mutagenic effects of FAA and MAA, the two metabolites that accumulate in hepatocytes deficient for FAH, were performed only in vitro (4). An advantage of our model is the ability to study the mutagenic effects of FAA in vivo. By analyzing mutations that occurred in the wild-type allele of Hgd in many different nodules, the spectrum of mutations caused by FAA (and possibly MAA) can be elucidated. As shown in Table 1, a wide variety of mutations were found in Hgd. This was surprising because FAA previously was predicted to be an alkylating agent, and this type of mutagen has been shown to cause mostly base substitutions (34). The mutation spectrum also differs from what is seen in the mouse liver because of spontaneous mutagenesis, where greater than 80% of the mutations are missense mutations (35).
In addition to the intragenic mutations found in Hgd, possibly up to 50% of the loss of heterozygosity in the nodules was caused by gene conversion or deletion of the Hgd gene, indicating that these events may be part of the mutation spectrum induced by FAA. However, our current data cannot differentiate between gene conversion and complete deletion of the Hgd gene. HT1 patients also have been reported to show increased chromosomal breakage (36), further supporting the possibility that FAA may cause more complex genetic changes including chromosomal damage or loss. Other mutagens, including the alkylating agent N-ethyl-N-nitrosourea, have not shown this propensity for inducing loss of heterozygosity (37).
Our model also could be used to study the mutation spectrum of FAA more extensively by breeding these mice with one of the transgenic mouse models that have been developed for easy detection of mutations, such as the Muta mouse (38). However, an advantage of our current model is the ability to detect larger deletions, whereas many of the transgenic mouse models cannot because of the limitations of bacteriophage packaging used in these models (39). Such limitations in other models may explain why larger deletions often have not been found with other alkylating agents.
The high probability for the development of liver cancer in HT1 (3) provides further proof of the potent effects of FAA and MAA. The frequency of nodule formation in the Hgd heterozygous mice indicates that the accumulation of these metabolites must be causing many mutations elsewhere in the genome of any FAH-deficient cells and certainly contributing to the formation of cancer in HT1 patients. A full analysis of the damage that results from the build-up of FAA and MAA in hepatocytes would allow for a better understanding of the pathophysiology of HT1 by using this in vivo mouse model.
Acknowledgments
We thank Xavier Montagutelli for the gift of the aku mice, Ching-Nan Ou for liver function analysis, Nancy Kennaway and Sesame Williams for amino acid analysis, and Angela Major and Billie Smith for their support with histological analysis. We also thank Mike Liskay for his review of the manuscript. This work was supported by National Institutes of Health Grant RO1-DK48252 to M.G.
Abbreviations
- HT1
hereditary tyrosinemia type I
- aku
alkaptonuria
- FAH
fumarylacetoacetate hydrolase
- HGD
homogentisic acid dioxygenase
- MAA
maleylacetoacetate
- FAA
fumarylacetoacetate
- NTBC
2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione
- HPD
4-hydroxyphenylpyruvate dioxygenase
- RT-PCR
reverse transcription–PCR
- H&E
hematoxylin/eosin
- ASO
allele-specific oligonucleotide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lindblad B, Lindstedt S, Steen G. Proc Natl Acad Sci USA. 1977;74:4641–4645. doi: 10.1073/pnas.74.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell G A, Grompe M, Lambert M, Tanguay R M. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W, Valle D, editors. Vol. 1. New York: McGraw–Hill; 1999. pp. 1077–1106. [Google Scholar]
- 3.Russo P, O’Regan S. Am J Hum Genet. 1990;47:317–324. [PMC free article] [PubMed] [Google Scholar]
- 4.Jorquera R, Tanguay R M. Biochem Biophys Res Commun. 1997;232:42–48. doi: 10.1006/bbrc.1997.6220. [DOI] [PubMed] [Google Scholar]
- 5.Lindstedt S, Holme E, Lock E A, Hjalmarson O, Strandvik B. Lancet. 1992;340:813–817. doi: 10.1016/0140-6736(92)92685-9. [DOI] [PubMed] [Google Scholar]
- 6.Holme E, Lindstedt S. J Inherited Metab Dis. 1998;21:507–517. doi: 10.1023/a:1005410820201. [DOI] [PubMed] [Google Scholar]
- 7.La Du B N. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W, Valle D, editors. Vol. 1. New York: McGraw–Hill; 1995. pp. 1371–1386. [Google Scholar]
- 8.Zannoni V G, Lomtevas N, Goldfinger S. Biochim Biophys Acta. 1969;177:94–105. doi: 10.1016/0304-4165(69)90068-3. [DOI] [PubMed] [Google Scholar]
- 9.Montagutelli X, Lalouette A, Coudé M, Kamoun P, Forest M, Guénet J L. Genomics. 1994;19:9–11. doi: 10.1006/geno.1994.1004. [DOI] [PubMed] [Google Scholar]
- 10.Manning K, Fernandez-Canon J M, Montagutelli X, Grompe M. Hum Mutat. 1999;13:171. doi: 10.1002/(SICI)1098-1004(1999)13:2<171::AID-HUMU15>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 11.Grompe M, Al-Dhalimy M, Finegold M, Ou C N, Burlingame T, Kennaway N G, Soriano P. Genes Dev. 1993;7:2298–2307. doi: 10.1101/gad.7.12a.2298. [DOI] [PubMed] [Google Scholar]
- 12.Grompe M, Lindstedt S, al-Dhalimy M, Kennaway N G, Papaconstantinou J, Torres-Ramos C A, Ou C N, Finegold M. Nat Genet. 1995;10:453–460. doi: 10.1038/ng0895-453. [DOI] [PubMed] [Google Scholar]
- 13.Edwards S W, Knox W E. In: Methods in Enzymology. Colowick S P, Kaplan N O, editors. Vol. 2. New York: Academic; 1955. pp. 292–295. [Google Scholar]
- 14.Fernández-Cañón J M, Peñalva M A. J Biol Chem. 1995;270:21199–21205. doi: 10.1074/jbc.270.36.21199. [DOI] [PubMed] [Google Scholar]
- 15.Sturman J A, Applegarth D A. In: Neuromethods. Boulton A A, Baker G B, Wood J D, editors. Vol. 3. Clifton, NJ: Humana; 1985. pp. 1–27. [Google Scholar]
- 16.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt S R, Gehrig A, Koehler M R, Schmid M, Muller C R, Kress W. Mamm Genome. 1997;8:168–171. doi: 10.1007/s003359900383. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D Y, Nozari G, Schold M, Conner B J, Wallace R B. DNA. 1989;8:135–142. doi: 10.1089/dna.1.1989.8.135. [DOI] [PubMed] [Google Scholar]
- 20.Miller S A, Dykes D D, Polesky H F. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhim J A, Sandgren E P, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1995;92:4942–4946. doi: 10.1073/pnas.92.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerdes J, Schwab U, Lemke H, Stein H. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 23.Fernández-Cañón J M, Granadino B, Beltrán-Valero C, Renedo M, Fernández-Ruiz E, Peñalva M A, Rodriguez de Córdoba S. Nat Genet. 1996;14:19–24. doi: 10.1038/ng0996-19. [DOI] [PubMed] [Google Scholar]
- 24.Endo F, Kubo S, Awata H, Kiwaki K, Katoh H, Kanegae Y, Saito I, Miyazaki J, Yamamoto T, Jakobs C, Hattori S, et al. J Biol Chem. 1997;272:24426–24432. doi: 10.1074/jbc.272.39.24426. [DOI] [PubMed] [Google Scholar]
- 25.Botstein D, Maurer R. Annu Rev Genet. 1982;16:61–83. doi: 10.1146/annurev.ge.16.120182.000425. [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Cañón J M, Peñalva M A. Proc Natl Acad Sci USA. 1995;92:9132–9136. doi: 10.1073/pnas.92.20.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kvittingen E A, Rootwelt H, Brandtzaeg P, Bergan A, Berger R. J Clin Invest. 1993;91:1816–1821. doi: 10.1172/JCI116393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirschhorn R, Yang D R, Puck J M, Huie M L, Jiang C K, Kurlandsky L E. Nat Genet. 1996;13:290–295. doi: 10.1038/ng0796-290. [DOI] [PubMed] [Google Scholar]
- 29.D’Andrea A D, Grompe M. Blood. 1997;90:1725–1736. [PubMed] [Google Scholar]
- 30.Ellis N A, Lennon D J, Proytcheva M, Alhadeff B, Henderson E E, German J. Am J Hum Genet. 1995;57:1019–1027. [PMC free article] [PubMed] [Google Scholar]
- 31.Jonkman M F, Scheffer H, Stulp R, Pas H H, Nijenhuis M, Heeres K, Owaribe K, Pulkkinen L, Uitto J. Cell. 1997;88:543–551. doi: 10.1016/s0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- 32.Ariga T, Yamada M, Sakiyama Y, Tatsuzawa O. Blood. 1998;92:699–701. [PubMed] [Google Scholar]
- 33.Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou C N, Finegold M, Grompe M. Nat Genet. 1996;12:266–373. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 34.Vogel E W, Nivard M J M. Mutat Res. 1994;305:13–32. doi: 10.1016/0027-5107(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 35.de Boer J G, Erfle H, Walsh D, Holcroft J, Provost J S, Rogers B, Tindall K R, Glickman B W. Environ Mol Mutagen. 1997;30:273–286. [PubMed] [Google Scholar]
- 36.Gilbert B E, Barness L A, Meisner L F. Pediatr Pathol. 1990;10:243–252. doi: 10.3109/15513819009067111. [DOI] [PubMed] [Google Scholar]
- 37.Wijnhoven S W, Van Sloun P P, Kool H J, Weeda G, Slater R, Lohman P H, van Zeeland A A, Vrieling H. Proc Natl Acad Sci USA. 1998;95:13759–13764. doi: 10.1073/pnas.95.23.13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gossen J A, de Leeuw W J, Vijg J. Mutat Res. 1994;307:451–459. doi: 10.1016/0027-5107(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 39.Vijg J, van Steeg H. Mutat Res. 1998;400:337–354. doi: 10.1016/s0027-5107(98)00030-x. [DOI] [PubMed] [Google Scholar]