Abstract
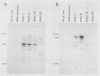
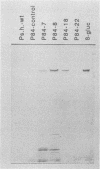
The plant-pathogenic fungus Pseudocercosporella herpotrichoides has been successfully transformed by using two different positive selection systems in combination with the Escherichia coli gusA gene. The selectable markers used in this study were the hygromycin B phosphotransferase gene (hph) from E. coli and the gene (bml) for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa. A lower transformation rate was obtained with the bml system than with the hph system. Conversely, cotransformation frequencies, as determined with medium plates containing the chromogenic substrate 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid, were higher with bml than with hph as the selectable marker. The hygromycin-resistant transformants were mitotically stable, and both the selectable gene and gusA were maintained through conidiation. The vector DNA was integrated into the genome, and the number and sites of insertion varied among transformants. Enzyme assays of mycelial extracts showed that beta-glucuronidase activity was highest in transformants with a high gusA copy number. Expression of gusA during growth of the fungus on plants was easily detectable and did not affect pathogenicity. These results form the basis for construction of a versatile and sensitive reporter gene system for P. herpotrichoides.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cullen D., Leong S. A., Wilson L. J., Henner D. J. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene. 1987;57(1):21–26. doi: 10.1016/0378-1119(87)90172-7. [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Klass M., Wolf N., Hirsh D. Expression of chimeric genes in Caenorhabditis elegans. J Mol Biol. 1987 Jan 5;193(1):41–46. doi: 10.1016/0022-2836(87)90624-3. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt P. J., Oliver R. P., Dingemanse M. A., Pouwels P. H., van den Hondel C. A. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56(1):117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- Roberts I. N., Oliver R. P., Punt P. J., van den Hondel C. A. Expression of the Escherichia coli beta-glucuronidase gene in industrial and phytopathogenic filamentous fungi. Curr Genet. 1989 Mar;15(3):177–180. doi: 10.1007/BF00435503. [DOI] [PubMed] [Google Scholar]
- Schmitz U. K., Lonsdale D. M., Jefferson R. A. Application of the beta-glucuronidase gene fusion system to Saccharomyces cerevisiae. Curr Genet. 1990 Mar;17(3):261–264. doi: 10.1007/BF00312618. [DOI] [PubMed] [Google Scholar]
- Wang J., Holden D. W., Leong S. A. Gene transfer system for the phytopathogenic fungus Ustilago maydis. Proc Natl Acad Sci U S A. 1988 Feb;85(3):865–869. doi: 10.1073/pnas.85.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]