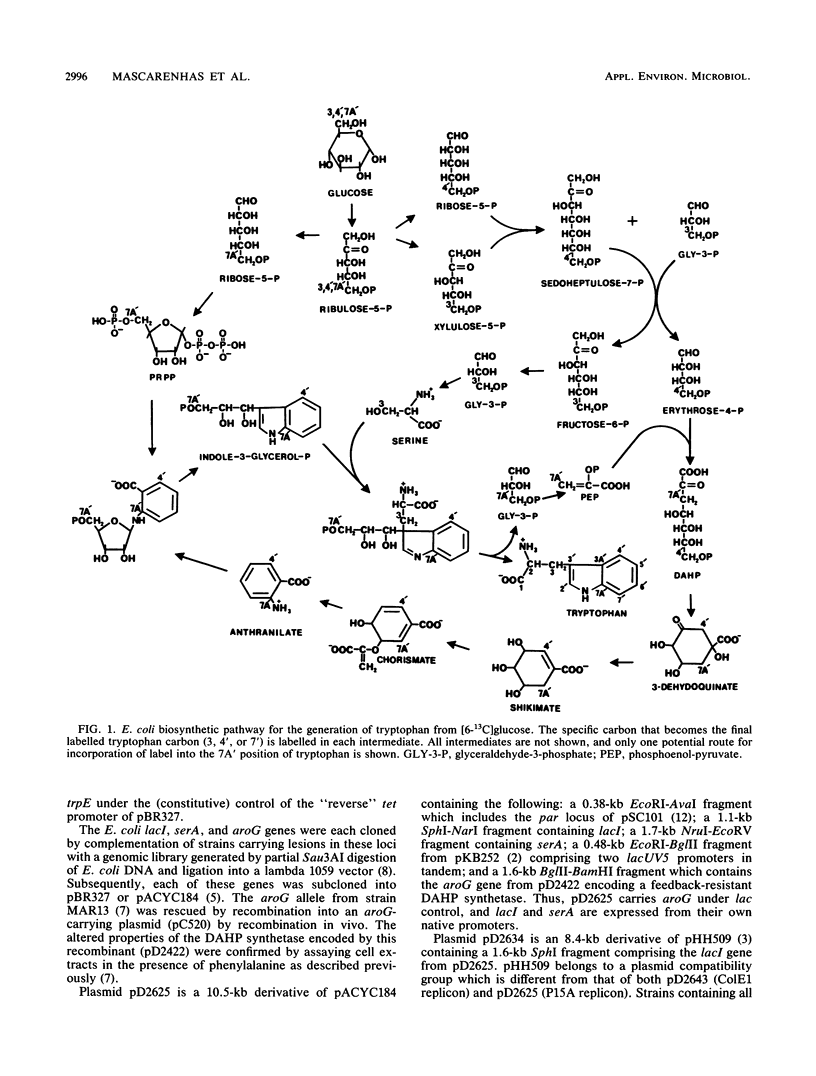
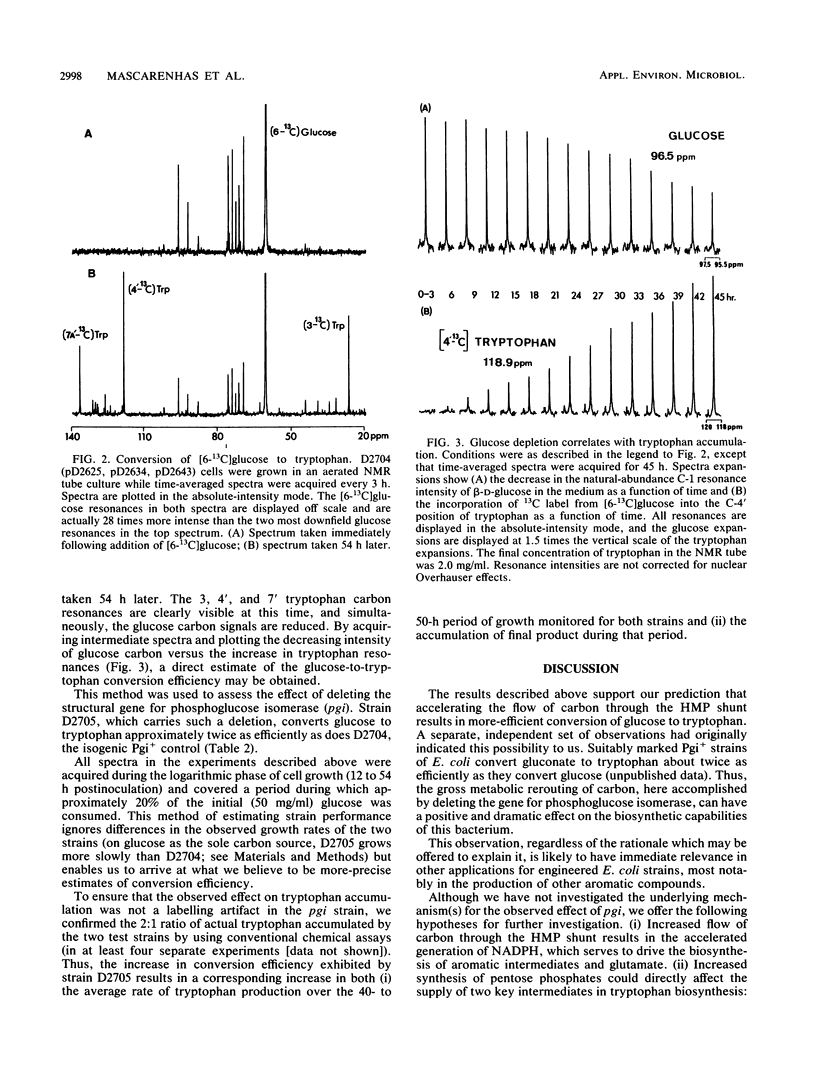
Abstract
Deletion of the structural gene for phosphoglucose isomerase (pgi) of Escherichia coli dramatically alters the path of glucose catabolism by diverting carbon into the hexose monophosphate shunt. The effect of this genetic alteration on the conversion of glucose to tryptophan by strains optimized for the biosynthesis of this amino acid was determined by using 13C-nuclear magnetic resonance spectroscopy in vivo. Pgi- strains converted glucose to tryptophan almost twice as efficiently as did their Pgi+ counterparts.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashworth D. J., Chen C. S., Mascarenhas D. Direct observation of tryptophan biosynthesis in Escherichia coli by carbon-13 nuclear magnetic resonance spectroscopy. Anal Chem. 1986 Mar;58(3):526–532. doi: 10.1021/ac00294a006. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Richards H., Datta N. Copy numbers of coexisting plasmids in Escherichia coli K-12. J Bacteriol. 1978 Sep;135(3):760–765. doi: 10.1128/jb.135.3.760-765.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedik M., Mascarenhas D., Campbell A. Probing cII and himA action at the integrase promoter pi of bacteriophage lambda. Gene. 1982 Oct;19(3):303–311. doi: 10.1016/0378-1119(82)90020-8. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Held W. A., Smith O. H. Mechanism of 3-methylanthranilic acid derepression of the tryptophan operon in Escherichia coli. J Bacteriol. 1970 Jan;101(1):209–217. doi: 10.1128/jb.101.1.209-217.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L., Cesareni G. Novel bacteriophage lambda cloning vector. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Mascarenhas D., Trueheart J., Benedik M., Campbell A. Retroregulation: control of integrase expression by the b2 region of bacteriophages lambda and 434. Virology. 1983 Jan 15;124(1):100–108. doi: 10.1016/0042-6822(83)90293-3. [DOI] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]


