Abstract
Images (i.e., figures) are important experimental evidence that are typically reported in bioscience full-text articles. Biologists need to access images to validate research facts and to formulate or to test novel research hypotheses. We propose to build a biological question answering system that provides experimental evidences as answers in response to biological questions. As a first step, we develop natural language processing techniques to identify sentences that summarize image content.
1. Introduction
Images (i.e., figures) are important experimental evidence that are typically reported in bioscience full-text articles. Biologists need to access images to validate research facts, and to formulate or to test novel research hypotheses. On the other hand, biologists live in an age of information explosion. As thousands of biomedical articles are published every day, systems that help biologists access efficiently images in literature would greatly facilitate biomedical research. Information retrieval systems (e.g., PubMed) typically return a list of documents in response to a user’s query. However, hundreds, thousands or even a larger number of documents can be retrieved; few biologists have time to read all of the retrieved documents. Additionally, many of the retrieved documents might not contain specific information that biologists need. For example, a biologist may specifically want to know the evidence of “X interacting with Y”, and he/she might have to navigate a large number of articles to identify the piece of information he/she needs. We are developing question answering techniques to automatically analyze thousands of documents and to extract answers in response to questions1-3. In the biology domain, we believe the first step to build a useful question answering system is to identify the text that summarize images that appear in full-text articles.
The motivations of this study come after our manual examination of more than one hundred questions posed by twelve experimental biologists. We found that many biological questions require experimental evidences as answers. In the following we listed five biological questions that require experimental evidences as answers:
Show me the evidences that the phosphatase domain and the C2 domain are responsible for membrane recruitment of PTEN.
Does bysl coexpress with myc?
Does ascorbic acid inhibit nitrosamines?
Give me all structures of E cahderin.
Show me ompa expression.
It is well-known that significant amount of experimental evidences are presented in the full-text body as images, including figures or tables, embedded within associated text. Because images are important experimental evidences, a question answering system may provide images as answers in response to biological questions.
However, images alone are frequently meaningless without their associated text (e.g. captions and other associated text). On the other hand, text alone without the images would deprive of the actual supporting experimental evidences.
To answer biological questions that require images as the answers, we propose to provide a short text that summarizes the content of the target images. Biologists can then access the actual images through the text. For example, to answer the question (1), we will provide the textual statement “Subcellular localization studies of PTEN transfected into HEK293T and HeLa cells indicated that targeting of PTEN to the plasma membrane is coupled with rapid degradation and that the phosphatase domain and the C2 domain are both necessary and sufficient for its membrane recruitment.” The corresponding image (i.e., image “C” in Figure 1) can be accessed directly from the preceding sentence with a hyperlink. To build such a question answering system, it is essential to develop natural language processing systems that automatically identify text that summarize images. We report natural language processing techniques to achieve such a mapping.
Figure 1.
: An example of a full-text biological article (pmid=12808147) in which the abstract sentences summarize the corresponding images (Arrows indicate the correspondences).
2. Text that Summarize Image Content
We randomly selected a total of ten Proceedings of the National Academy of Sciences (PNAS) full-text articles; we manually examined the associated text (e.g., abstract sentences, and image captions and other associated text that appear in the full-text body). As an example, the following shows the associated text of “Table 1” in Figure 1.
Abstract sentences
Our in vitro surface plasmon resonance measurements using immobilized vesicles showed that both the phosphatase domain and the C2 domain, but not the C-terminal tail, are involved in electrostatic membrane binding of PTEN. Furthermore, the phosphorylation-mimicking mutation on the C-terminal tail of PTEN caused an 80-fold reduction in its membrane affinity, mainly by slowing the membrane-association step.
Image caption
Binding parameters for PTEN constructs determined from SPR analysis.
Other associated text
The sample cell contains the sensor surface coated with the vesicles indicated in Table 1.
Further measurements using mixed vesicles of POPC/POPS (8:2), which roughly approximate the lipid composition of the inner plasma membrane of mammalian cells to which PTEN is targeted, showed that both the full-length PTEN and the C2 domain had higher affinity (in terms of Kd) for these anionic vesicles (see Table 1).
The affinity for anionic vesicles was mainly attributed to nonspecific electrostatic interactions, because they did not distinguish POPC/POPS (8:2) from POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) (8:2) vesicles (Table 1) and the affinity was reduced greatly by 0.5 M KCl [Kd = (2.2 ± 0.4) x 10−7 M; ka = (9.5 ± 1.2) x 103 M−1·s−1; kd = (2.1 ± 0.3) x 10−3· s−1].
As shown in Table 1, Δ 1−185 and Δ353–403 had 31- and 1.5-fold lower affinity for POPC/POPS (8:2) vesicles than WT, respectively. This suggests that the phosphatase domain is important for the membrane binding of PTEN, whereas the C-terminal tail plays practically no role in membrane binding.
As shown in Table 1, PTEN had 2-fold higher affinity for the plasma membrane mimic than for POPC/POPS (8:2) vesicles, presumably because the former has a higher anionic lipid content.
As shown in the example above, the abstract sentences are the best to summarize image content. Other associated text typically describe only experimental procedures and do not include indications and conclusions of an experiment. Additionally, image content are typically scattered across other associated text, and therefore, it is difficult to identify a succinct summary from the other associated text.
3. An Annotated Corpus in Which Abstract Sentences Link to Images that Appear in the Full-Text Documents
We hypothesize that images reported in a full-text article can be summarized by sentences in the abstract. To test this hypothesis, we randomly selected a total of 329 biological articles that are recently published in leading biological journals Cell (104), EMBO (72), Journal of Biological Chemistry (92), and Proceedings of the National Academy of Sciences (PNAS) (61). For each article, we emailed the corresponding author to ask him or her to identify abstract sentences that summarize the image content in that article.
A total of 119 biologists from 19 countries participated voluntarily the annotation to identify abstract sentences that summarize figures or tables in their 114 articles (39 Cells, 29 EMBO, 30 Journal of Biological Chemistry, and 16 PNAS), a collection that is 34.7% of the total articles we requested. The responding biologists included the corresponding authors to whom we had sent emails, as well as the first authors of the articles to whom the corresponding authors had forwarded our emails. None of the biologists were compensated.
This collection of 114 full-text articles incorporates 742 images and 826 abstract sentences. The average number of images per document is 6.5±1.5 and the average number of sentences per abstract is 7.2±1.9. Our data show that 87.9% images correspond to abstract sentences and 66.5% abstract sentences correspond to images; those statistics have empirically validated our hypothesis that image content can be summarized by abstract sentences. This collection of 114 annotated articles was then used as the corpus to evaluate our natural language processing approaches that automatically identify abstract sentences that summarize image content in the full-text articles.
4. NLP Approaches that Link Abstract Sentences to Images
Linking abstract sentences to images is a task of linking the abstract sentences to other associated text of the images. Our study is built upon two assumptions. The first is that image content consistently corresponds to its associated text. The second is that there are strong lexical similarities between the text associated with each image and the corresponding sentence(s) in the abstract. We empirically evaluated both assumptions.
We deployed hierarchical clustering techniques4 to cluster abstract sentences and images based on the lexical similarities. The details of the study are reported elsewhere5. In the following sections, we describe a few systems we explored including the clustering algorithms and feature selections.
4.1. Clustering Algorithms
Hierarchical clustering algorithms are widely used in many other areas including biological sequence alignment6, gene expression analysis7, and topic detection8. The algorithm starts with the set of text that includes abstract sentences or image captions. Each sentence or image caption represents a document that needs to be clustered. The algorithm identifies pairwise document similarities based on the TF*IDF weighted cosine similarity. It then merges the two documents with the highest similarity into one cluster. It then re-evaluates pairs of documents/clusters; two clusters can be merged if the average similarity across all pairs of documents within the two clusters exceeds a predefined threshold. When multiple clusters can be merged at any time, the pair of clusters with the highest similarity is always preferred.
4.2. Features, Weights, Clustering Strategies
We explored bag-of-words as the learning features. We explored three different methods to obtain the IDF value for each word feature: 1) IDF(abstract+caption): the IDF values were calculated from the pool of abstract sentences and image captions; 2) IDF(full-text): the IDF values were calculated from all sentences in the full-text article; and 3) IDF(abstract)::IDF(caption): we obtained two sets of IDF values. For word features that appear in abstracts, the IDF values were calculated from the abstract sentences; for words that appear in image captions, the IDF values were calculated from the image captions.
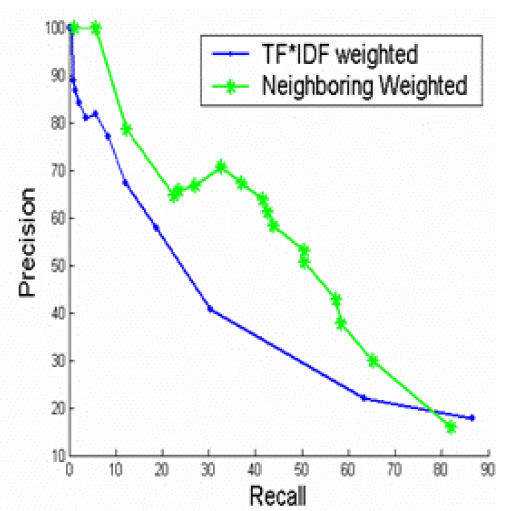
We also explored the position features. The positions of abstract sentences that correspond to images seem to have a “neighboring effect”. The chance that two abstract sentences link to an image decreases when the distance between two abstract sentences increases.
To integrate “neighboring effect” into our existing hierarchical clustering algorithms, we modified the TF*IDF weighted cosine similarity with neighboring weights. Assuming that we consider an abstract sentence or an image caption as a document, the TF*IDF weighted cosine similarity for a pair of document i and j is Sim(i,j), we integrated the “neighboring effect” and the final similarity W(i,j) is:
If i and j are both abstract sentences, Ti=Tj=total number of abstract sentences; and Pi and Pj represents the positions of sentences i and j in the abstract.
If i and j are both image captions, Ti=Tj=total number of images that appear in a full-text article; and Pi and Pj represents the positions of images i and j in the full-text article.
If i and j are an abstract sentence and an image caption, respectively, Ti=total number of abstract sentences and Tj=total number of images that appear in a full-text article; and Pi and Pj represent the positions of abstract sentence i and image j.
Finally, we have explored three clustering strategies; namely, per-image, per-abstract sentence, and mix.
Per-image clusters each image caption with all abstract sentences. The image is assigned to (an) abstract sentence(s) if they belong to the same cluster. This method values features in abstract sentences more than image captions because the decision that an image belongs to (a) sentence(s) depends upon the features from all abstract sentences and the examined image caption. The features from other image captions will not play a role for the clustering.
Per-abstract-sentence takes each abstract sentence and clusters it with all image captions that appear in a full-text article. Images are assigned to the sentence if they belong to the same cluster. This method values features in image captions higher than the features in abstract sentences because the decision that an abstract sentence belongs to image(s) depends upon the features from the image captions and the examined abstract sentence. The features from other abstract sentences will not play a role for the clustering.
Mix clusters all image captions with all abstract sentences. This method treats features in abstract sentences and image captions equally.
5. Results and Discussion
Figure 2 shows that the “local” IDFs, or the IDFs calculated from the abstract sentences and image captions yield much better performance than the “global” IDFs, or the IDFs that are calculated from the full-text article. The results are not surprising because “local” IDFs reflect more on the characteristics of the full-text article in study than the “global” IDFs. Figure 3 show that Per-image out-performs the other two strategies. The results suggest that features in abstract sentences are more useful than features in caption for the task of clustering. Figure 4 shows that “neighboring weighted” approach has a significant enhancement over the TF*IDF weighted approach. The recall ranges from 0% to 88%, while precision ranges from 100% to 8%. When the recall is 33%, the precision of “neighboring weighted” approach increases to 72% from the original 38%, which corresponds to a 34% absolute increase. The results strongly indicate the importance of “neighboring effect” or positions as additional features. When the precision is 100%, the recall is 4.6%. A high precision is the key to success for this application. Many successful natural language processing systems achieved high precisions with a cost of lower recall9.
Figure 2.
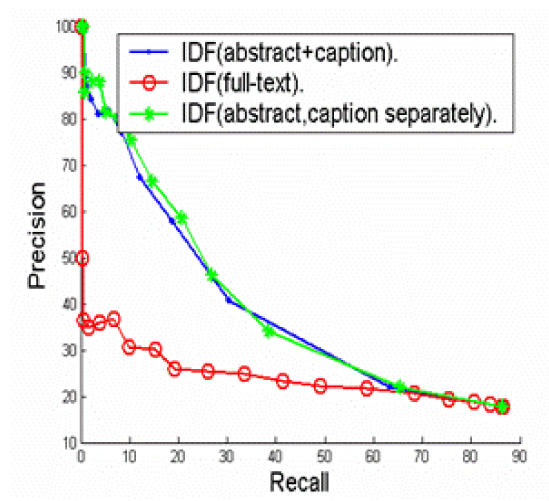
Figure 3.
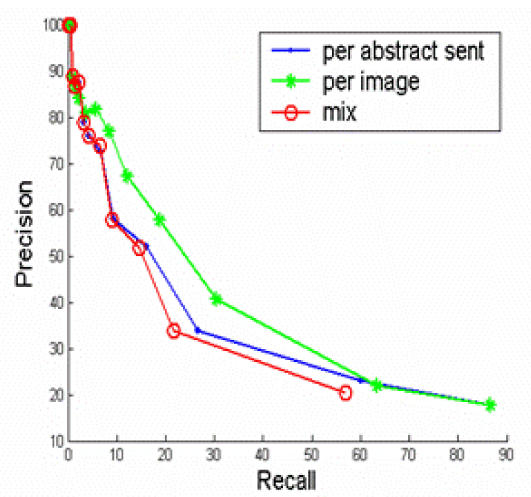
Figure 4.
6. Future Work
Our long term goal is to integrate the body of this work into our larger question answering system3. Additionally, we believe that we have room to enhance the performance for linking abstract sentences to images. We believe that “orderly effect”, which is that the position order of image pairs (i.e., one image appears ahead of the other image) reflects the position order of abstract sentences, may be a useful feature to further enhance the task for linking abstract sentences to images. To test this hypothesis, we examined the statistics in our data set. Excluding images that have not been annotated to any sentences, we found a total of 1,640 image pairs. Of those only 433 pairs are reversed, and therefore a total of 1,207 or 73.6% image pairs appear with the same order of their corresponding sentences. To integrate “ordering effect” into existing algorithm, we may apply first a high-precision clustering algorithm to identify the first match between abstract sentences to images, and then cluster the preceding sentences with preceding abstracts and the following sentences with following abstracts to capture the next matches.
7. Related Work
Research and systems have been developed in experimental data retrieval. Tulipano and colleagues10 defined an image taxonomy (e.g., “imaging instruments” and “imageable probes” are two children concepts of “imaging entity”) and linked the image taxonomy to the Gene Ontology11. BioImage is a web-based object-oriented image database12. The SLIP system identifies images that depict protein subcellular locations13. PDB is a database for biological macromolecules and their relationships to sequence, function and disease14. Captions were explored for image classification in newswire images15. However, none of the existing systems incorporates the natural language processing techniques (e.g., summarization and question answering) to map images to text.
Acknowledgement
The author thanks Dr. Weiqing Wang for collecting some annotations from biologists, Minsuk Lee for implementing the hierarchical clustering systems, and Philip Payne for valuable comments. The author was supported by JDRF 6-2005-835. Any opinions, findings, or recommendations are those of the authors and do not necessarily reflect JDRF’s views.
9. References
- 1.Yu H, Sable C. Nineteenth International Joint Conference on Artificial Intelligence on Knowledge and Reasoning for Answering Questions; 2005. [Google Scholar]
- 2.Yu H, Sable C, Zhu H. AAAI Workshop on Qusetion Answering in Restricted Domains; 2005. [Google Scholar]
- 3.Yu H, et al. Beyond information retrieval--Biomedical question answering. AMIA. 2006 [Google Scholar]
- 4.Lee M, Wang W, Yu H. Exploring supervised and unsupervised methods to detect topics in Biomedical text. BMC Bioinformatics. 2006;7:140. doi: 10.1186/1471-2105-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Lee M. 14th Annual International Conference on Intelligent Systems for Molecular Biology; Brazil. 2006. [Google Scholar]
- 6.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrero J, Valencia A, Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics. 2001;17:126–136. doi: 10.1093/bioinformatics/17.2.126. [DOI] [PubMed] [Google Scholar]
- 8.Lee M, Wang W, Yu H. Exploring supervised and unsupervised approaches to detect topics in biomedical text. BMC Bioinformatics, Accepted. 2006 doi: 10.1186/1471-2105-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman C, Kra P, Yu H, Krauthammer M, Rzhetsky A. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics. 2001;17(Suppl 1):S74-82. doi: 10.1093/bioinformatics/17.suppl_1.s74. [DOI] [PubMed] [Google Scholar]
- 10.Tulipano PK, Millar WS, Cimino JJ. Linking molecular imaging terminology to the gene ontology (GO) Pac Symp Biocomput. 2003:613–623. doi: 10.1142/9789812776303_0057. [DOI] [PubMed] [Google Scholar]
- 11.Consortium GO. The Gene Ontology (GO) databases and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catton C, Shotton D. 3rd E-BioSci/Oriel Annual Workshop. Hinxton; Cambridge, UK: 2004. [Google Scholar]
- 13.Murphy RF, Velliste M, Yao JGP. IEEE International Symposium on Bio-Informatics and Biomedical Engineering (BIBE) 119-128. 2001. [Google Scholar]
- 14.Sussman JL, et al. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1998;54:1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 15.Sable C, Hatzivassiloglou V. Text-based approaches for non-topical image categorization. International Journal on Digital Libraries. 1999 [Google Scholar]



