Abstract
Imaging plays an important role in characterizing tumors. Knowledge inferred from imaging data has the potential to improve disease management dramatically, but physicians lack a tool to easily interpret and manipulate the data. A probabilistic disease model, such as a Bayesian belief network, may be used to quantitatively model relationships found in the data. In this paper, a framework is presented that enables visual querying of an underlying disease model via a query-by-example paradigm. Users draw graphical metaphors to visually represent features in their query. The structure and parameters specified within the model guide the user through query formulation by determining when a user may draw a particular metaphor. Spatial and geometrical features are automatically extracted from the query diagram and used to instantiate the probabilistic model in order to answer the query. An implementation is described in the context of managing patients with brain tumors.
Introduction
Advances in medical imaging have significantly increased the amount of clinical data that is available to physicians. Medical images provide an objective source for documenting a patient’s illness and encourage the practice of evidence-based medicine [1]. In particular, imaging plays a critical role in guiding the treatment of oncology patients: primary and indicator lesions are visually tracked over time to gauge response to a treatment. Additionally, changes in contrast, texture, and boundaries that characterize a tumor can provide further correlation to prognosis [2]. Medical imaging has the potential to dramatically improve disease management, but physicians need a system that organizes the data, enables quantitative analysis on the data, and provides the user with a visual environment for interacting with such data.
The goal of this work is to develop a system that assists users in utilizing the available knowledge to answer three basic clinical questions: diagnosis (e.g., what type of tumor does the patient have?), prognosis (e.g., what is the time to survival for a 28-year old patient presented with a particular lesion type?), and treatment (e.g., what combination of therapies will maximize survival and quality of life?). A key component in this system is the visual interface with which users pose queries to a probabilistic disease model. Interfaces employing various querying paradigms (e.g., tabular, natural language) have been developed to address the limitations of traditional database formalisms, such as structured query language (SQL). From the perspective of working with images, a more natural approach is to pose queries using graphical metaphors [3], which are visual representations of variables or parameters specified in the disease model. The system then automatically translates the arrangement of graphical metaphors into a visual query. A disadvantage of existing visual query interfaces is that users are presented with a potentially large set of graphical metaphors but receive little or no guidance on how to effectively utilize these metaphors to construct a query. An adaptive palette is introduced that utilizes the structure and parameters within the underlying Bayesian belief network (BBN) to filter irrelevant metaphors. This approach reduces the potential visual clutter associated with presenting the entire set of metaphors simultaneously and assists users in selecting which metaphor is best suited for a particular query. Unlike previous work [4] in developing intelligent user interfaces, the adaptive palette is designed to be unobtrusive to the user and is bounded to a domain specified by the Bayesian belief network.
Background
Several tools such as Prefuse [5] and H3 [6], which visualize generic graphs, or VisNet [7], which specializes in BBNs, have addressed the issue of model complexity by exploiting graphical features such as temporal order, color, size, proximity, and animation to emphasize relationships and the relative importance of individual variables. Other tools such as GeNIe [8], HUGIN [9], and SamIAm [10] combine model visualization with a mechanism to perform different computational analysis on the network. Although these systems provide the ability for exploration and analysis of network variables, their interfaces are not inherently conducive for posing clinical questions. Promedas [11] is a probabilistic decision support system designed for clinicians, but it limits the user to diagnostic queries and requires the user to answer long lists of questions.
Another design consideration is generating a suitable set of graphical metaphors to represent image features. These metaphors must be developed such that the graphical representation is universally recognizable and unambiguous. [12] provides a taxonomy of medical data visualizations and describes a method to evaluate the design of icons and graphical primitive. Additionally, works on query-by-example [13, 14] paradigms used in content-based image retrieval have demonstrated that when working with images, visual querying provides a highly accurate and intuitive method for interacting with multimedia data. Having the user draw a query, rather than input an actual image of a tumor (e.g., ASSERT [15]), provides more flexibility in posing queries and the ability to experiment with different parameters. This paper illustrates how the combination of a query-by-example paradigm with the use of graphical metaphors and a probabilistic model facilitates the exploration and explanation of disease behavior and improve patient outcome.
System Framework
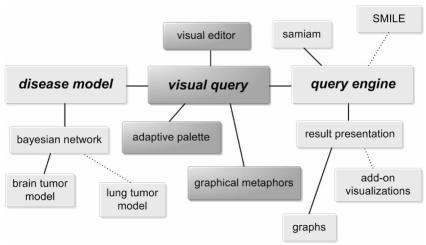
The overall system is comprised of three components, as depicted in Fig. 1: (1) a disease model specifies the relationships among variables; (2) an adaptive visual query interface based on a query-by-example paradigm enables users to pose a query pictorially; and (3) an engine instantiates the model, executes the query, and presents the results. To illustrate a possible application of the framework, an implementation is described using a brain tumor Bayesian belief network.
Figure 1.
A diagrammatic representation of the overall system. Darkened boxes represent components described in this paper. Dotted lines depict how individual components may be added or customized.
A. Probabilistic Disease Model
Central to this framework is the probabilistic disease model that quantitatively describes the relationships among features (evidence variables) extracted from the data. The framework is designed to accommodate a variety of probabilistic models (e.g., Markov, BBN), but for this instance, the Bayesian belief network of a brain tumor is used. The benefit of using a BBN over another representation (e.g., rule-based) is its ability to model uncertainty, accommodate large amount of information efficiently, and provide quantitative explanations to a variety of clinical questions [16].
A Bayesian belief network is a model that reflects the probabilistic relationships among evidence variables. Formally depicted as a directed acyclic graph, the nodes represent evidence variables and the edges represent a relationship that one node has on another. Associated with each node is a conditional probability table (CPT) that quantifies the effects that the parent node has on its children nodes. In disease models, the conditional probability distributions may be populated with data from actual patient cases and/or published scientific literature [16,17]. For this implementation, an imaging-based disease model has been developed from an existing neuro-oncology brain tumor patient database with associated radiology and medical reports. Currently fifteen imaging variables such as necrosis, edema, and rim contrast; and non-imaging variables such as age, sex, and Karnofsky Performance Score are incorporated into the model. A set of 143 patients was initially used to compute the conditional probabilities for an imaging-based network topology. As more data is collected, the network may be expanded to accommodate additional relevant variables, and the CPTs refined.
Each evidence variable in the model is assigned to a semantic group, which divides the entire set of variables into smaller, related collections. Three initial semantic groups are defined for this model: patient, anatomical level, and disease. The user starts a query by defining the most general parameters, such as patient demographics. Next, the anatomical level is defined by selecting a particular image slice in a labeled atlas. Then, through the iterative process of selecting and modifying metaphors, the user creates a pictorial representation of the disease state. Additional groups (e.g., treatment, genetic) may be specified depending on the number and types of variables in a model. The purpose of semantic grouping is to divide a complex disease model into smaller sub-models, which are then further parsed using methodology discussed in the next section.
B. Visual Editor
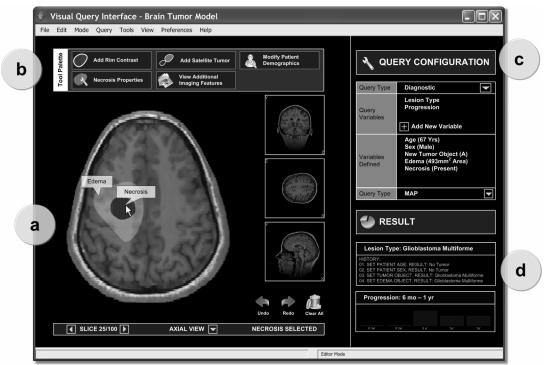
The visual editor (Fig. 2a) is the system’s front-end through which users interact with the disease model. The primary component of the editor is an image viewer that displays slices from a three-dimensional digitized atlas of the region of interest. The user first selects an orientation (axial, sagittal, or coronal) and then chooses a representative image from a set of slices to depict the location of the tumor. The anatomical metadata associated with the atlas (e.g., location and boundary of the ventricles) are used to provide contextual information regarding the existence of structures found within the selected slice [18]. The editor provides the user with functionality to select, draw, and customize graphical metaphors. The intent is to allow users to compose and overlay metaphors that pictorially represent a set of image findings on a single, representative slice of the atlas.
Figure 2.
A screen capture of the prototype visual query interface with a brain tumor model. The left side features the visual editor (a) and adaptive palette (b) where users create and pose the query while the right side includes panels for query configuration (c) and result presentation (d).
Adaptive palette
An adaptive palette (Fig. 2b) presents available metaphors above the editor. Metaphors are presented based on context; as the user selects structures (e.g., white matter) or metaphors (e.g., edema metaphor) in the visual editor, related metaphors are presented in the palette while unrelated metaphors are removed. The determination of whether a metaphor is related or unrelated is influenced by the relationships among variables defined in the disease model. Each metaphor is mapped to a corresponding variable in the model. Variables that are connected by an arc in the model are related. Therefore, selecting a particular metaphor will cause its related metaphors to be presented. Moreover, states listed in a variable’s CPT determine which attributes may be assigned to that metaphor. When the rim contrast metaphor is selected, the user is prompted to define whether the border is thick or thin. This adaptive presentation of relevant metaphors not only simplifies the process of creating a query by reducing visual clutter but also enforces logical rules regarding the order that metaphors are selected to formulate a query. For instance, in a magnetic resonance (MR) image, contrast enhancement, if present, appears around certain image features of a tumor, such as a cyst or necrosis. Therefore, the option to add a rim contrast metaphor is only available when a cyst or necrosis metaphor is already present in the query.
Query types
The editor permits the user to classify the query into four types: freeform, diagnostic, prognostic, and treatment-related queries. The choice of query type influences how results are presented. Results from diagnostic queries, for instance, emphasize what the most probable lesion type is given the patient’s characteristics. A freeform query, on the other hand, would allow the user to select and display a combination of results from the three other query types.
C. Query Engine & Result Presentation
Executing queries against the BBN is accomplished by using the engine implemented in [10]. Currently, three classes of queries are supported. A probability of evidence query finds the probability that a certain instantiation of variables would occur. The maximum a posteriori (MAP) query finds the most likely scenario or explanation of a subset of network variables. A specialization of MAP is the most probable explanation (MPE) query. MPE finds the most likely values of every variable given some evidence and is computationally less demanding than a MAP query. For the predefined query types, a MAP query is used to return a predefined subset of relevant variables. For freeform queries, either a MPE or probability of evidence query is executed depending on the configuration that the user selects.
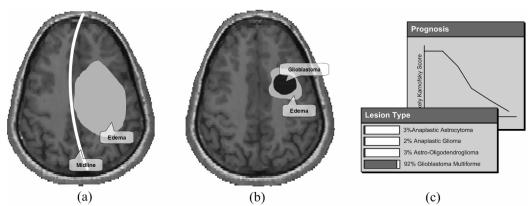
A wide range of clinically-relevant questions may be posed to the disease model using a combination of metaphors. A possible freeform query would be, “what are the states of all other evidence variables in the model, given symptoms of severe edema with noticeable mass effect and midline shift?” (Fig. 3a) This question is an example of a MPE query where the most probable states of all the variables within the network are calculated. If the user was interested in finding a treatment for the patient with these characteristics, he could choose to display the most common treatment along with the most probable patient outcome.
Figure 3.
(a) A sample diagnostic query given a patient with severe edema and noticeable mass effect. (b) A sample prognostic query given an area of mild edema and defined region of glioblastoma. (c) Example of how results from a diagnostic and prognostic query may be presented.
Additionally, the user may pose questions that relate to prognosis of a patient given a set of characteristics. For example, a prognostic query would be, “what is the most probable value of the Karnofsky Performance Score given that a 29 year old female has glioblastoma, with MR findings showing a mild edema?” (Fig. 3b) In this query, three metaphors (lesion, lesion type, and edema) are used. First, the user draws a lesion metaphor on the brain. Then, the user modifies the metaphor’s attributes, defining it as glioblastoma. Finally, the user sketches a small area of edema surrounding the lesion. This query is executed as a MAP query, and the most probable value of the Karnofsky Performance Score is returned.
Depending on the initial query type selected, the results are presented using various representations (Fig. 3c). The system also permits a user to alter a particular attribute while keeping all others constant and displays a comparison of how the results change between the original and new values. Visualizing the influence that one factor has on the overall disease allows physicians to better understand the impact of a particular treatment on patient outcome.
Discussion
Rapid developments in imaging technology are making possible the acquisition of physical and quantitative data that thoroughly and accurately characterizes a disease. Integrating this knowledge into daily patient care would improve a physician’s ability to diagnose and treat a patient. The interface presented in this paper describes a methodology that enables physicians to interact with a complex disease model by creating a pictorial representation of their desired query through the use of graphical metaphors. By varying the usage and attributes of these metaphors, a wide variety of queries such as diagnostic, prognostic, and treatment-type questions may be posed. Furthermore, the visual query interface introduces a methodology for presenting graphical metaphors based on context: depending on how the user interacts with existing structures and metaphors, only related and valid metaphors are presented. Past research [19] demonstrates that users perform well using conceptual interfaces. Though performance varies with user groups, this system has been designed with the feedback of physicians, and a pilot study of this system shows similar promising results.
Although this paper describes the system in the domain of neuro-oncology, the system is extensible to other domains given a probabilistic model, an engine capable of querying the model, and any necessary contextual information (e.g., labeled anatomical atlas). Furthermore, the semantic groups presented are only a sampling of the possible classifications. One approach would be to use a comprehensive medical ontology such as Unified Medical Language System (UMLS) to automatically classify evidence variables.
Potential future directions include extending the interface to enable queries across a period of time. An example would be calculating the time to survival for a patient given that his tumor has doubled in size over a period of half a year. Additionally, a full evaluation of the user interface and sensitivity of the model will be completed.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Suzie El-Saden and Dr. Hooshang Kangarloo for their feedback. This research was supported in part by the NLM Training Grant LM007356.
References
- 1.Kaplan B, Shaw NT. People, organizational, and social issues: Evaluation as an exemplar. In: Haux R, Kulikowski C, editors. Yearbook of Medical Informatics. Stuttgart: Shattauer; 2002. pp. 91–102. [PubMed] [Google Scholar]
- 2.Pope WB, Sayre A, Perlina A, Villablanca JP, Mischel P, Cloughesy T. Magnetic resonance imaging correlates of survival in patients with high grade gliomas. Am. J. Neuroradiology. 2005;26:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 3.Marcus A. CHI 98: Conf. Human Factors Computing Systems. Los Angeles, CA: 1998. Apr-23. Metaphor design for user interfaces; pp. 129–130. [Google Scholar]
- 4.Horvitz E, Breese J, Heckerman D, Hovel D, Rommelse K. The Lumiere project: Bayesian user modeling for inferring the goals and needs of software users. Proc. 14th Conf. on UAI; 1998 July; Madison, WI. pp. 256–265. [Google Scholar]
- 5.Heer J, Card SK, Landay JA. Prefuse: A toolkit for interactive information visualization. CHI 05: Conf. Human Factors Computing Systems; 2004 Apr 2–7; Portland, OR. pp. 421–430. [Google Scholar]
- 6.Munzner T. H3: Laying out large directed graphs in 3D hyperbolic space. Proc. IEEE Symp. Information Visualization. 1997:2–10. [Google Scholar]
- 7.Zapata-Rivera JD, Greer JE. Inspecting and visualizing distributed Bayesian student models. Proc. 5th Int. Conf. Intelligent Tutoring Systems. 2000:544–553. [Google Scholar]
- 8.Druzdzel MJ. GeNIe: A development environment for graphical decision-analytic models. Proc. Ann. Symp. AMIA. 1999:1206. [Google Scholar]
- 9.Andersen SK, Olesen KG, Jensen FV, Jensen F. HUGIN-a Shell for Building Belief Universes for Expert Systems. Proc. IJCAI. 1989:1080–1085. [Google Scholar]
- 10.Sensitivity Analysis, Modeling, Inference, and More (SamIam) main web site. [Last accessed February 20, 2006.]. http://reasoning.cs.ucla.edu/samiam/
- 11.Promedas. [Last accessed February 20, 2006.]. http://www.promedas.nl/
- 12.Starren J, Johnson SB. An object-oriented taxonomy of medical data presentations. 2000 Jan-Feb;7(1):1–20. doi: 10.1136/jamia.2000.0070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasso G, Marsiglia HR, Pigatto F, Basilicata A, Gargiulo M, Abate AF, et al. A visual query-by-example image database for chest CT images: Potential role as a decision and educational support tool for radiologists. J Digital Imaging. 2005 Mar;18(1):78–84. doi: 10.1007/s10278-004-1025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egenhofer MJ. Query processing in spatial-query-by-sketch. J. Visual Languages and Computing. 1997;8(4):403–424. [Google Scholar]
- 15.Shyu CR, Brodley CE, Kak AC, Kosaka A, Aisen AM, Broderick LS. ASSERT: A physician-in-the-loop content-based retrieval system for HCRT image databases. Comp. Vis. Image. Und. 1999;75(12):111–132. [Google Scholar]
- 16.Burnside E. Bayesian networks: computer-assisted diagnosis support in radiology. J. Acad. Rad. 2005;12(4):422–430. doi: 10.1016/j.acra.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 17.Herckeman DE, Horvitz EJ, Nathwani BN. Toward normative expert systems. Part I. The pathfinder project. Methods Inform. Med. 1992;31:90–105. [PubMed] [Google Scholar]
- 18.Sinha U, Taira RK, Kangarloo H. Structure localization in brain images - Application to relevant image selection. Proc. Ann. Symp. AMIA. 2001:622–627. [PMC free article] [PubMed] [Google Scholar]
- 19.Siau KL, Chan HC, Wei KK. Effects of query complexity and learning on novice user query performance with conceptual and logical database interfaces. In IEEE Trans. Sys. Man. & Cybernetics. 2004;34(2):276–281. [Google Scholar]