Abstract
Scientists in small research labs need more robust tools than spreadsheets to manage their data. However, no suitable laboratory information management systems (LIMS) are readily available; they are either too costly or too complex. We have therefore developed Seedpod, a model driven LIMS that allows users to create an integrated model of a LIMS without programming. Seedpod then automatically produces a relational database from the model, and dynamically generates a web-based graphical user interface. Our goal is to make LIMS easier to use by decreasing development time and cost, thereby allowing researchers to focus on producing and collecting data.
1. Introduction
Effective data management continues to be a bottleneck in the life sciences research process [1]. Most of today’s scientific and clinical researchers in small-sized labs use spreadsheets to manage their data. Spreadsheets are easy to use, readily available, and allow users to start collecting data quickly. Scientists like spreadsheets for data management because they can focus on what data to store in the cells of the grids without knowing how to store the data. They can organize data in whatever manner they want, and can perform calculations on the fly, all without the aid of a programmer. However, spreadsheets are inadequate for dealing with increasingly large datasets, complex data relationships, multimedia data, and collaborative research groups. Thus, scientists need more robust solutions, in the form of laboratory information management systems (LIMS).
Although LIMS are effective in the pharmaceutical and biotech industries [2], most are too costly and not readily available for small research labs. Many labs across the country develop and use their own lab-specific LIMS, but building a custom LIMS for each lab generally requires considerable programming in order to specify how the data are stored and accessed.
Our long-term goal is to create a LIMS-building toolkit that retains the ease-of-use of the spreadsheet while gaining the robustness of a LIMS. As one step towards that goal we have developed Seedpod, a model driven LIMS building toolkit that allows a user to create a model that specifies not only what the types of data and their relationships are, but also what the behavior of the application should be, all with minimal or no programming. Thus, by eliminating or greatly reducing the time needed to program how the system stores and accesses data, the model driven-approach provides a foundation on which to build the tools that will let individual users create their own LIMS through the graphical specification of models.
In the current Seedpod implementation, the model is created by an informaticist using the frame-based Protégé knowledge representation system [3] (Section 2.2). Even though there is a learning curve for using Protégé, but the reduced need for custom programming greatly reduces the overall development time over a non-model based approach. We developed a formal method that automatically generates a relational database schema from the Protégé model (Section 2.3). The Seedpod server then interprets the model and dynamically generates a web-based user interface (Section 2.4). Scientists can browse and manage their data stored in the relational database through this web-based user interface. The following sections describe these steps in more detail.
2. Seedpod System Description
The driving application for the development of Seedpod is an information management system to help our Human Brain Project collaborator at the University of Washington, Dr. George Ojemann. Dr. Ojemann’s lab studies the functional anatomy of speech and speech memory. In some of his studies a technique called single unit recording (SUR) is used to record a large amount of high temporal resolution neuronal signal data during open brain surgery. Scientists in the lab correlate the electrical signal data with behavioral and other data in order to find the meaning behind neural activities.
The Ojemann lab is a small one, and has used spreadsheets to record data because of their ease of use. The lab stores numeric or string data in spreadsheets, but manages multimedia data, such as the neuronal firing patterns, in a separate file system. The file system uses a complicated naming convention, which is managed manually. Data version control, coordinated data entry, and data sharing are challenging due to the lack of a centralized management system that can be accessed through the Internet. In addition, searching the data requires meticulous hand trimming and picking of datasets from multiple Excel sheets. Such searches are becoming increasingly untenable as the number of SUR studies increase. We have built Seedpod partly in response to these problems.
2.1. Seedpod’s General Architecture
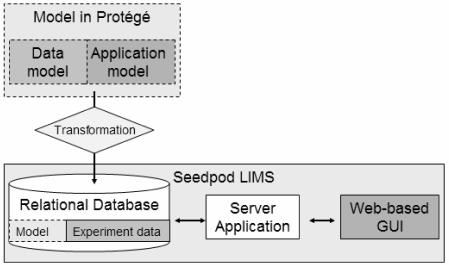
There are two major components in Seedpod: the model and the LIMS application engine. The model is an integrated representation of a LIMS (Figure 1). It includes a domain-specific data model describing the entities and relationships that the scientist wants to manage. It also includes an application model describing properties that allow the scientist to customize the look and feel of the LIMS web-based user interface.
Figure 1.
Seedpod architecture: the Protégé model (top) and the web-based LIMS application (bottom).
The LIMS application engine has a server application, a backend relational database, and a web-based graphical user interface (GUI). Seedpod automatically transforms the Protégé model into a relational schema for the relational database. The database stores the experiment data and the model. The server application queries the database regarding the model, retrieves and stores the experiment data, and creates dynamic web pages for users based on the look and feel specified in the application model. The Protégé model and LIMS application are not linked in real time (i.e., they can change and evolve independent of each other).
2.2 LIMS Model
The first step in implementing a Seedpod-based project is to create a LIMS model using Protégé. Protégé is a frame-based knowledge management tool (http://protege.stanford.edu/). We choose to use Protégé primarily because of its expressivity in model representation [3].
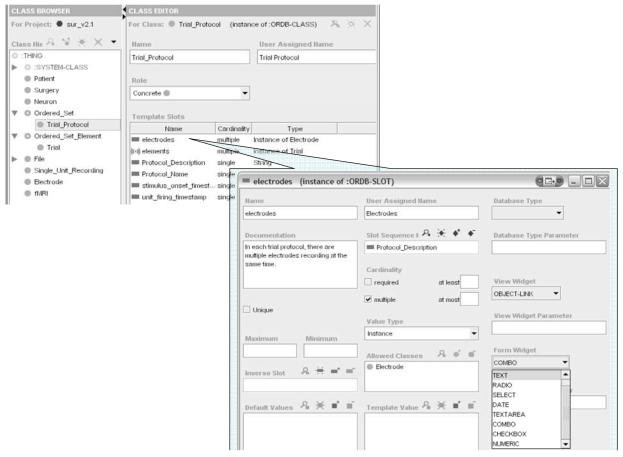
A Protégé model consists of a set of named classes. Figure 2 shows a Protégé screen shot of class Trial_Protocol in the Ojemann SUR model. The class hierarchy of the model is shown on the left. Each class instance is associated with a set of template-slots, which are properties that are propagated to its instances and children classes. The value of a template-slot can be a primitive type such as String, Integer, Boolean, etc. Protocol_Description in Figure 2 is an example of a slot with a String type value. A template-slot can also be a relationship type, which has class instances as its values. The template-slot electrodes in Figure 2 is such a relationship slot. This slot describes a one-to-many relationship from a Trial_Protocol instance to multiple Electrode instances. The valid values for the slot are class instances of the Electrode class.
Figure 2.
Sample screen of a Protégé model for a single unit recording experiment. In the upper left panel template-slots listed to the right are slots for the highlighted class, Trial_Protocol. The output database schema for this class is shown in Listing 1. Details of the template-slot, electrode, are shown in the lower right panel.
This Protégé model differs from an entity-relational (ER) model because it is object-oriented. It does not require the user to understand normalization in data modeling. It allows users to easily describe common yet complex biological data structures such as inheritance, object relationships, and many-to-many relationships. The model designer does not need to be relational database experts.
Additionally, the Protégé model in Seedpod includes the application model as well as the data model. For example, we can extend the standard template-slot definition to allow users to customize the look and feel of the LIMS application. The expanded window of the electrodes slot in Figure 2 shows a slot definition that we have extended to include “Database Type,” user interface “View Widget,” “Form Widget,” etc. Protégé makes this extension possible by allowing the modeler to extend the standard slot definition.
2.3 Model Transformation and Data Storage
As described in Section 2.1, each Seedpod application stores its experiment data in a relational database for efficient storage and retrieval. It is important to note that Seedpod does not use a generic database schema to handle all labs. Any given lab will have its own specific Protégé model, which is transformed into a specific relational schema. Instead of transforming Protégé models to schema manually (ad hoc), we developed a generalized method that performs this transformation automatically [4].
The formal definitions of the relational and frame-based models provided the inspiration for the following rules:
Class transformations
A class, C, is transformed to a relational table or a view (or both):
T1 If C is a concrete class, then create a table with name C_table and add a primary key attribute ID.
T2 If C has subclasses, (is non-leaf) then create a view, C_union, that is defined by selecting the union of C_table and all of its subclasses’ tables.
Slot transformations
A slot, S, of a class, C, is transformed depending on the slot's value type and cardinality.
T3 If the range of S is primitive (i.e., String, Integer, Float, Boolean or Symbol), and has cardinality of 1, then create an attribute Attr_S for table C_table, and give it the corresponding relational database primitive type.
T4 If the range of S is instances of class B, and has cardinality 1, then create a foreign key attribute, FK_S (in table C_table) that references the ID attribute in table B_table.
T5 If S has cardinality multiple, create a new association table, Assoc_S. Add a foreign key, FK_S (in table Assoc_S) that references the ID attribute in table C_table. Create an attribute Attr_S for S in Assoc_S according to single cardinality rules T3 or T4.
Additional rules dealing with multiple cardinality slots, relational slots, class inheritance, etc. are beyond the scope of this paper (see [4] and [5]).
The transformation method is implemented in a JAVA program. It takes a Protégé model file as input, and outputs the relational schema in SQL statements in a text file. Listing 1 shows the result of transforming class Trial_Protocol from the SUR model (described in Section 2.2 and Figure 2) to its database schema. The LIMS developer loads the SQL command file (e.g., Listing 1) into a relational database engine to create the database. We use a PostgreSQL database in the Seedpod prototype.
Listing 1.
A sample relational schema output shows SQL statements from the transformation for table Trial_Protocol, and a view for its superclass Ordered_Set.
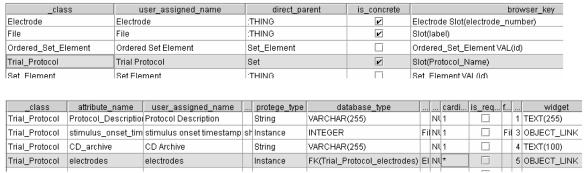
In addition to the data tables (like those shown in Listing 1) the database also stores the LIMS model in two tables, one table for classes (Figure 3 top) and the other for slots (Figure 3 bottom). Figure 3 shows screenshots of the tables with our previous examples, class Trial_Protocol and slot electrodes, highlighted. The Seedpod engine queries these two tables for information about the model while dynamically generating the web application.
Figure 3.
The model is stored in the relational database as two Seedpod application tables called _class (top) and _attribute (bottom). Table _class (top) shows an example of the class Trial_Protocol. The table _attribute (bottom) shows slot electrodes of Trial_Protocol. The Seedpod server application queries these two tables for information about the LIMS model.
2.4 Seedpod Application
The Seedpod server application is generic; it is not specific for any model. After a LIMS model is completed and loaded into the database, the Seedpod server connects to the database server to deploy the web application and start collecting data. The server application queries the model in the database (Figure 3) to dynamically generate the web-based user interface for the scientists.
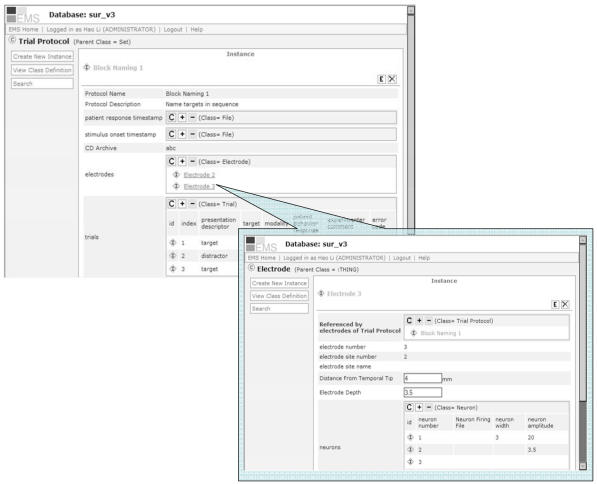
The scientist can browse and manage data in the relational database through this user interface. For example, the web page for an instance of a Protégé class in the SUR model, Trial_Protocol, displays the template-slots and their values (Figure 4). The display name of the instance is specified in the model as the slot value of Protocol_Name (also visible in Figure 3 (top) as Slot(Protocol_Name) under the “browser_key” column). The application renders the slot values using information from the model such as slot layout sequences and form field widgets. Figure 3b (bottom) shows that slot electrodes uses widget OBJECT_LINK. In Figure 4, the values for electrodes slot are rendered as two URL links. Clicking on one of the links takes users to the corresponding web page displaying Electrode instances. See [5] for a live demo of Seedpod.
Figure 4.
Screenshots of Seedpod’s web-based graphical user interface. The page on top shows an instance of the Trial_Protocol class. The page displays its template-slots and their values. Users can browse data related to this instance through a URL link created for its relationship slot, electrodes.
Seedpod’s server application is implemented using Tomcat and JAVA. For each class in the model, the application dispatches by reflection the appropriate JAVA class implementation based on the class’s identity in the model. For example, if a user wants to view an instance of a Trial_Protocol, the engine tries to dispatch the Trial_Protocol JAVA class implementation to handle the request. However, if the Trial_Protocol JAVA class does not exist, the engine tries to find Trial_Protocol’s parent class, Order_Set_Element’s JAVA class. It continues until an implementation is found. Most of the classes in the SUR model are handled by the default class. This mechanism allows the LIMS developer to extend the server application by implementing class plug-ins.
3. Discussion
The motivation for this work is the scientists’ need for tools beyond the spreadsheet for data management. The model driven approach taken by Seedpod moves in this direction by removing or reducing the need to program how data are stored and accessed, allowing the developer to concentrate instead on specifying what the data look like.
The only existing LIMS toolkit we are aware of that takes a similar model driven approach is Teranode’s XDA, which offers an integrated workflow data modeling environment in addition to laboratory process automation for data collection [6]. However, the Protégé modeling environment is richer than that used by Teranode. Not only does Protégé make it easier to model complex data relationships, it also allows us the freedom to extend the modeling constructs, capturing additional metadata about workflow, interaction among data elements, GUI customization, and access to controlled terminologies that are represented as Protégé ontologies.
Currently, Seedpod requires an expert Protégé user to create the model. In our future work, we will develop an integrated graphical design environment, on top of Protégé, which will make it easier for scientists to model their data and experiment workflow. We will also test the generalizability of Seedpod by deploying the system in other scientific research domains. It is our ultimate goal to allow scientists to create and deploy their own LIMS using Seedpod without the help of informaticists.
Acknowledgments
4. Acknowledgement
Supported by NIH Human Brain Project Grant RO1 MH/DC02310, and National Library of Medicine Training Grant program number: T15-LM07442.
5. References
- 1.Lacroix Z, Critchlow T. Bioinformatics: Managing Scientific Data. Morgan Kaufmann; Jul, 2003. [Google Scholar]
- 2.Paszko C, Turner E. Laboratory Information Management Systems. 2. New York: Marcel Dekker; 2002. [Google Scholar]
- 3.Gennari J, Musen MA, Fergerson RW, Grosso WE, Crubézy M, Eriksson H, et al. The evolution of Protégé: an environment for knowledge-based system development. International Journal of Human-Computer Studies. 2003;58(1):89–123. [Google Scholar]
- 4.Gennari J, Mork P, Li H. Knowledge Transformations between Frame Systems and RDB Systems. In proceedings of the K-CAP05 Conference; pp. 197–198. [Google Scholar]
- 5.Seedpod Project URL: http://sig.biostr.washington.edu/projects/seedpod;2006
- 6.Teranode. 2006. Home Page. http://teranode.com/