Abstract
A fundamental aspect of the biogenesis and function of eukaryotic messenger RNA is the quality control systems that recognize and degrade non-functional mRNAs. Eukaryotic mRNAs where translation termination occurs too soon (nonsense-mediated decay)1 or fails to occur (non-stop decay)2 are rapidly degraded. We show that yeast mRNAs with stalls in translation elongation are recognized and targeted for endonucleolytic cleavage, referred to as ‘no-go decay’. The cleavage triggered by no-go decay is dependent on translation and involves Dom34p and Hbs1p. Dom34p and Hbs1p are similar to the translation termination factors eRF1 and eRF3 (refs 3, 4), indicating that these proteins might function in recognizing the stalled ribosome and triggering endonucleolytic cleavage. No-go decay provides a mechanism for clearing the cell of stalled translation elongation complexes, which could occur as a result of damaged mRNAs or ribosomes, or as a mechanism of post-transcriptional control.
To determine the effect of a delay in translation elongation on mRNA decay, we inserted a stable stem-loop, previously shown to inhibit translation elongation5, into the coding region of a yeast PGK1 reporter construct6 to give the PGK1-SL reporter. We observed that the PGK1-SL mRNA decays faster than the control PGK1 mRNA in wild-type yeast cells (Supplementary Fig. 1), suggesting that a ribosomal stall accelerated degradation of the mRNA.
In yeast, the two major pathways of mRNA turnover initiate with deadenylation of the 3′ -polyadenosine (poly(A)) tail7. Deadenylation is followed either by decapping and 5′ →3′ degradation, or 3′ →5′ degradation7, of the mRNA. The PGK1-SL mRNA showed accelerated decay in both a dcp2Δ strain, which lacks decapping activity8, and a ski7Δ strain, which is defective in the 3′ → 5′ degradation pathway2 (Supplementary Fig. 1). Moreover, the PGK1-SL mRNA showed the same instability in an upf1Δ strain, which is defective for the nonsense-mediated decay (NMD) pathway1 (Supplementary Fig. 1). Thus, the PGK1-SL mRNA shows accelerated degradation in a manner independent of known degradation pathways, suggesting that it is targeted for degradation by a novel mechanism, possibly involving cleavage by an endonuclease.
If the PGK1-SL mRNA is subject to cleavage by an endonuclease, two mRNA fragments should be produced, a 5′ fragment expected to be a substrate for the 3′ → 5′ degradation complex termed the exosome, and a 3′ fragment that would be uncapped and predicted to be a substrate for Xrn1p, the major 5′ →3′ exonuclease. Given this, we examined the PGK1-SL mRNA in strains defective in either exosome-mediated decay or Xrn1p function, using probes specific to the 5′ and 3′ portions of the mRNA.
Strains defective in cytoplasmic 3′ →5′ degradation accumulated a fragment of the PGK1-SL mRNA detected with a probe 5′ of the stem-loop (Fig. 1a). This 5′ fragment accumulated in strains defective in 3′ →5′ decay for any of three reasons: first, the loss of any component of the heterotrimeric Ski complex of Ski2, Ski3 and Ski8 proteins (ski2Δ, ski3Δ or ski8Δ), which is required for cytoplasmic exosome function9; second, the loss of Ski7p (ski7Δ), which couples the Ski complex to the exosome; and last, a point mutation in an exosome subunit (ski4-1)10 that blocks the interaction of the exosome with Ski7p (data not shown). This mRNA fragment was about 1,200 nucleotides long, which is the distance from the cap to the site of the stem-loop. These observations suggest that the PGK1-SL mRNA is subject to an endonucleolytic cleavage, with the 5′ mRNA fragment being degraded by the cytoplasmic exosome.
Figure 1. Stalling of elongating ribosomes leads to endonucleolytic cleavage of the PGK1-SL reporter transcript.
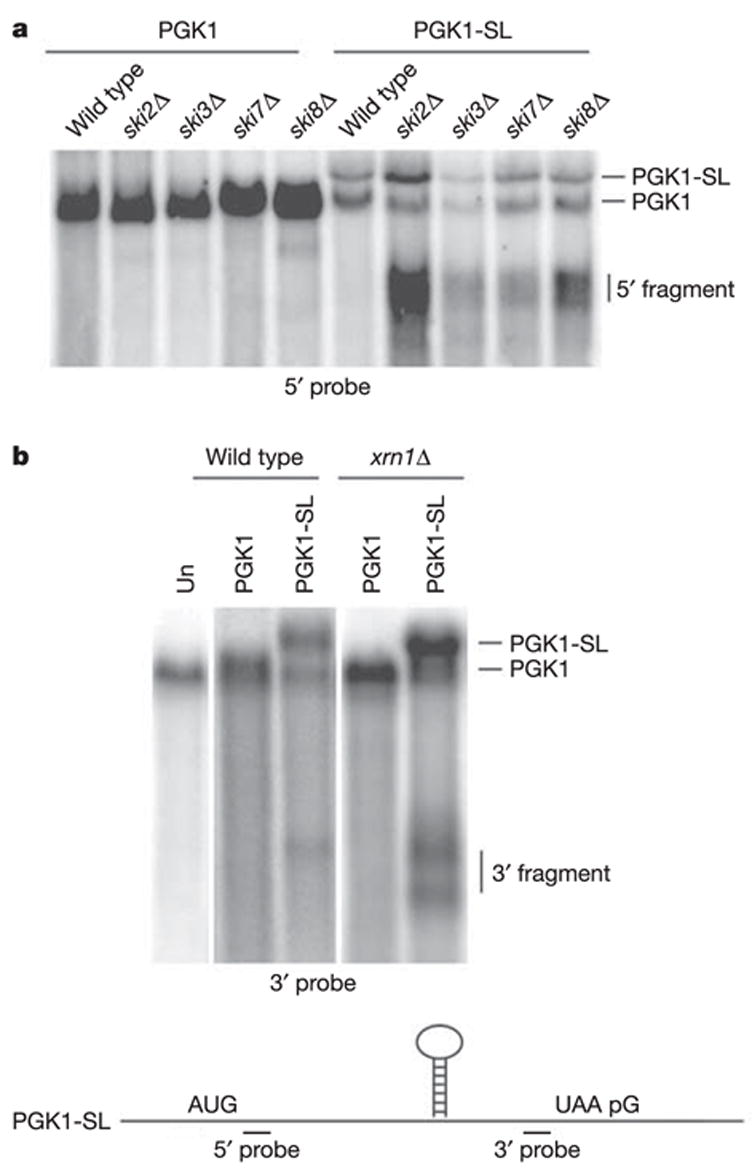
Northern analysis of steady-state PGK1 and PGK1-SL reporter mRNA in wild-type strains and strains mutant either in 3′ → 5′ exosome activity (a) that include ski2Δ, ski3Δ, ski7Δ and ski8Δ, or a mutant in 5′ → 3′ exonucleolytic activity, xrn1Δ (b). The untransformed strain (Un) is shown in b. Northern analysis was performed with probes specific for mRNA regions 5′ (a) or 3′ (b) of the stall site, as illustrated at the bottom.
Additional evidence that the PGK1-SL mRNA is subject to endonucleolytic cleavage is that a probe specific for the 3′ end of the PGK1-SL reporter detected 3′ decay intermediates in the xrn1Δ strain (Fig. 1b). The larger fragment is about 600 nucleotides long and corresponds to the distance from the stem-loop to the 3′ end of the mRNA. The smaller fragment is an mRNA fragment that has subsequently been trimmed at the 3′ end to a poly(G) tract present in the PGK1 3′ untranslated region, which blocks further 3′ → 5′ degradation10 (data not shown, and see below). The roughly 600-nucleotide 3′ mRNA fragment is present at lower levels in wild-type cells and mutant ski strains (Fig. 1a, and Supplementary Fig. 2a), which indicates that the inserted stem-loop structure can act as a block to Xrn1p (see below). We cannot formally determine whether the mRNA fragment accumulating in the wild-type strain arises by decapping and 5′ → 3′ degradation by Xrn1p, or by endonucleolytic cleavage. However, the mRNA fragment detected in the xrn1Δ strain must be produced by endonucleolytic cleavage, arguing that at least some of the 3′ decay product seen in the wild-type strain is produced by endonucleolytic cleavage. A probe specific to the 5′ end of the PGK1-SL mRNA did not identify any fragment that specifically accumulated in the xrn1Δ strain (Supplementary Fig. 2b).
The accumulation of a 3′ mRNA fragment in xrn1Δ strains, and a 5′ mRNA fragment in skiΔ mutants, indicated that the PGK1-SL mRNA was endonucleolytically cleaved followed by exonuclease digestion of the mRNA fragments. We refer to this surveillance mechanism as ‘no-go decay’ (NGD). Insertion of the stem-loop into the coding region of the MFA2 mRNA (MFA2-SL) also identified 5′ and 3′ decay intermediates in the ski7Δ and xrn1Δ mutants, respectively (Supplementary Fig. 3). Mapping of the 5′ and 3′ fragments for both the MFA2-SL and PGK1-SL transcripts indicates that the cleavage occurs in the vicinity of the stem-loop (Supplementary Fig. 3), although there is heterogeneity indicating that the cleavage occurs at multiple sites near the ribosomal stall site and/or that there is trimming by exonucleases from the original cleavage site(s). In addition, transcriptional pulse–chase experiments and transcriptional induction experiments indicate that the mRNA fragments are produced after the full-length mRNA has been synthesized, which is consistent with a precursor–product relationship between the full-length mRNA and the mRNA decay product (Supplementary Fig. 4, and data not shown).
Two observations indicate that deadenylation is not required to occur before endonucleolytic cleavage in NGD (Supplementary Fig. 5): first, in a transcriptional induction experiment, we observed that the 3′ mRNA fragment of the MFA2-SL mRNA was polyadenylated when first produced; and second, loss of the major mRNA deadenylase, Ccr4p, does not prevent NGD.
Other stalls in translation, including a pseudoknot, rare codons and premature translation termination codons, also induce cleavage events at low temperatures, although at greatly reduced levels (Fig. 2a). This observation supports the argument that the trigger for NGD might be a delay in translation elongation.
Figure 2. NGD is initiated by endonucleolytic cleavage of mRNA with various ribosomal stalls.
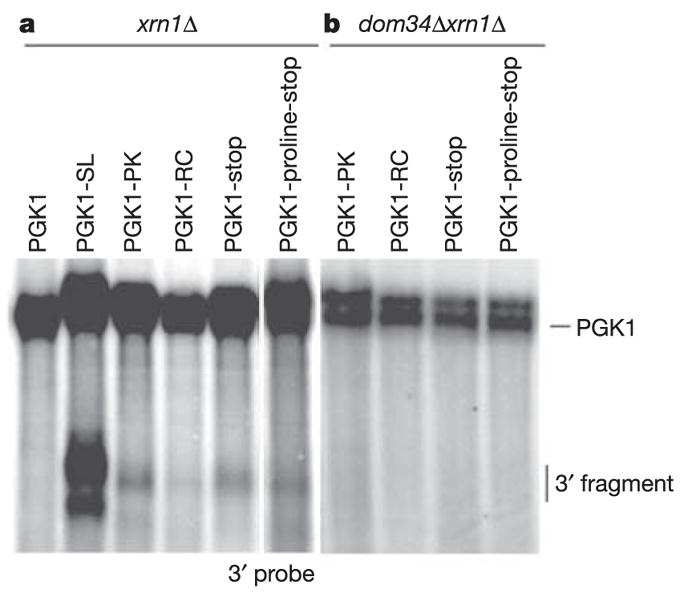
Steady-state cultures of xrn1Δ (a) and dom34Δxrn1Δ (b) strains with PGK1, PGK1-SL, PGK1-PK (pseudoknot), PGK1-RC (rare codons), PGK1-stop (stop codon) and PGK1-proline-stop (proline-stop codon) reporters were grown at low temperature (16 8C). Northern analysis of steady-state reporter mRNAs was performed with a probe specific for mRNA regions 3′ of the stall sites.
If stalls in translation elongation trigger NGD, ribosomes should be required to reach the stem-loop to trigger the cleavage event. To test this prediction, we prevented ribosomes from reaching the stall site by blocking ribosome scanning to the AUG by inserting a stem-loop in the 5 ′ untranslated region of the PGK1-SL mRNA11 (SL-PGK1-SL). This block to translation initiation prevented the accumulation of the 5′ and 3′ decay intermediates in the ski7Δ and xrn1Δ strains (Fig. 3; SL-PGK1-SL). In addition, the accumulation of the 5′ and 3′ fragments in ski7Δ and xrn1Δ strains was abolished when a premature stop codon was inserted in the PGK1-SL mRNA at codon 22 (PGK1-PTC-SL), such that ribosomes terminated translation before the ribosomal stall site12 (Fig. 3; PGK1-PTC-SL). Because the PGK1-PTC-SL mRNA is a substrate for NMD, its levels are quite low. To verify that these low levels of mRNA did not obscure the accumulation of the NGD products, we also analysed this mRNA in upf1Δski7Δ and upf1Δxrn1Δ strains, in which NMD is inactivated and mRNA levels are higher. Again, no 5′ or 3′ cleavage products accumulated (Fig. 3). Thus, the endonucleolytic cleavage we detect requires ribosomes to translate to the stem-loop, suggesting that ribosome stalling leads to cleavage.
Figure 3. NGD is dependent on translation by ribosomes.
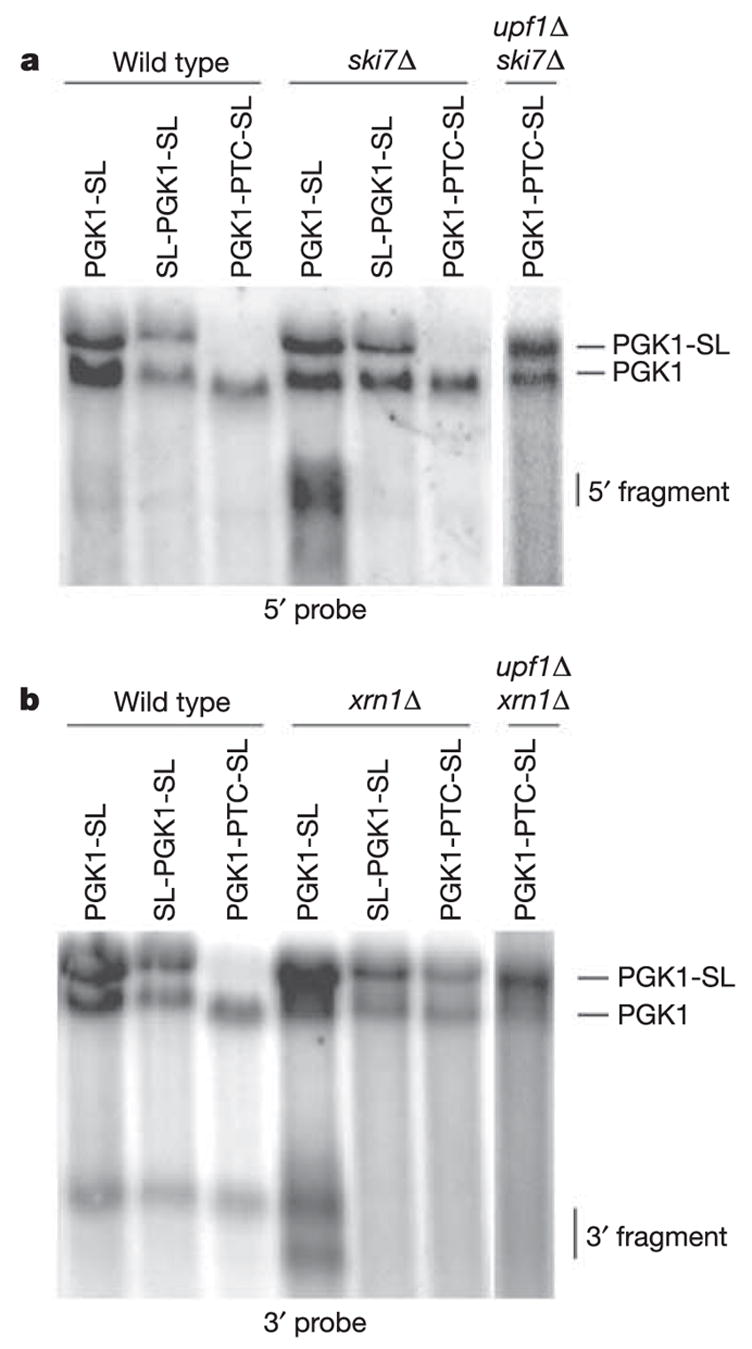
Northern analysis of steady-state PGK1-SL, SL-PGK1-SL (pRP1252) and PGK1-PTC-SL (pRP1253) reporter transcripts in wild-type, ski7Δ, upf1Δ ski7Δ (a) and xrn1Δ, upf1Δ xrn1Δ (b) mutant strains. Northern analysis was performed with probes specific for mRNA regions 5′ (a) and 3′ (b) of the stall site.
Because NGD works on stalled ribosomes, we proposed that Hbs1p and Dom34p, two conserved and interacting proteins with similarity to translation termination factors, might function in NGD. Hbs1p is likely to interact with the ribosome because it is a member of the family of GTPases consisting of eEF1A, which delivers the transfer RNA to the A site of the ribosome13, eRF3, which functions in translation termination14, and Ski7p, which is thought to interact with an empty A site when the ribosome reaches the 3′ end of a mRNA2. Moreover, Dom34p binds Hbs1p (ref. 15) and is related to eRF1, which has a three-dimensional structure similar to a tRNA and functions with eRF3 during translation termination16.
To test whether Hbs1p and/or Dom34p function in NGD, we determined whether the loss of Hbs1p or Dom34p affected the accumulation of NGD decay products from the PGK1-SL mRNA easily seen in ski7Δ and xrn1Δ strains. In both dom34Δski7Δ and dom34Δxrn1Δ strains the mRNA fragments generated by NGD were no longer observed (Fig. 4), indicating that Dom34p is required for NGD. Similar results were seen with other translational stalls (Fig. 2b).
Figure 4. Dom34p and Hbs1p affect NGD.

Northern analysis of steady-state PGK1-SL reporter mRNA in wild-type and mutant strains with probes specific for mRNA regions 5′ (a) and 3′ (b) of the stall site.
Hbs1p also has a function in NGD, because accumulation of 5′ fragments from the PGK1-SL mRNA observed in skiΔ mutants is substantially reduced in an hbs1Δski7Δor hbs1Δski2Δstrain (Fig. 4a). Surprisingly, and in contrast to the dom34Δxrn1Δ strain, the hbs1Δxrn1Δ strain still showed the accumulation of the 3′ fragment. This observation could be explained by either of two related possibilities. First, in an xrn1Δ strain, Ski7p might be able to substitute for the role of Hbs1p, because Ski7p and Hbs1p have recently diverged17. Second, Hbs1p might be required for NGD only when the cytoplasmic exosome is compromised, as seen in the hbs1Δski7Δ and hbs1Δski2Δ strains, perhaps because of a loss of Ski7p function in those mutant strains.
The above observations suggest that Dom34p recognizes stalled ribosomes, possibly in conjunction with Hbs1p, leading to endonucleolytic cleavage of the mRNA. It remains to be determined whether Dom34p is the endonuclease, activates a nuclease activity within the ribosome or recruits an endonuclease. Moreover, the identification of Dom34p and Hbs1p as mediators of NGD adds to the family of related proteins thought to modulate different events at the A site of the ribosome.
NGD provides a mechanism to release stalled, or non-functional, ribosomes and degrade damaged mRNA. An interesting possibility is that NGD has been adapted for the regulation of specific mRNAs, and previous examples of regulated ribosome-dependent cleavage of mRNAs in eukaryotes18,19 may be due to an inhibition of translation elongation, which then activates NGD. Consistent with this model is the observation that the sequences triggering endonucleolytic cleavage of the CGS1 mRNA in Arabidopsis have been recently shown to mediate ribosome stalling20. Finally, elongation pauses can also trigger endonucleolytic cleavage in prokaryotes21–24, indicating that mRNA surveillance based on translation elongation rates is broadly conserved.
METHODS
The stem-loop structure (SL) was constructed by using a synthetic oligonucleotide with the following sequence and is similar to one used in an earlier study5: 5′ -GAT ATC CCG TGG AGG GGC GCG TGG TGG CGG CTG CAG CCG CCA CCA CGC GCC CCT CCA CGG GAT ATC-3′. This was introduced into the pJD375 construct25 between Sal1 and BamH1 sites in the multiple cloning regions to give pRP1250. To generate the reporters used in this study, 275-base-pair fragments containing the SL was amplified by polymerase chain reaction with the use of primers containing Xba1 ends and ligated into the Xba1 site of wild-type PGK1 (pRP469) to give the PGK1-SL construct (pRP1251). Insertion of other translational stalls in the PGK1 mRNA, including the pseudoknot26, rare codons27, stop codon and proline-stop codon28 was done in the same manner, generating PGK1-PK(pRP1285), PGK1-RC(pRP1286), PGK1-stop(pRP1287) and PGK1-proline-stop(pRP1288) reporters. Primer sequences are available from the authors on request. The reporters used for the translational dependence of NGD were constructed by introduction of the Xba1-generated fragment containing the SL into pRP543 (ref. 11) and pRP609 (ref. 12) to give pRP1252 and pRP1253, respectively. All PGK1 constructs are under the control of the Gal1 upstream activation sequence (UAS). Yeast strains are detailed in Supplementary Information.
All RNA analyses were performed as described previously12. All experiments were performed at 30 °C in galactose-containing medium with mid-log cultures except for those in the experiments in Fig. 2, which were grown at 16 °C. For the PGK1 reporter mRNAs, the oligonucleotide oRP132 was used for detection of the 5′ fragment and the oligonucleotide oRP70 for detection of the 3′ fragment. Loading corrections for quantification were determined by hybridization with oRP100, an oligonucleotide directed to SCR1 RNA, a stable RNA polymerase III transcript.
Supplementary Material
Acknowledgments
We thank P. Farabaugh for the pJD375 plasmid; A. van Hoof for strains; and members of the Parker laboratory, especially K. Baker and C. Decker, for discussions. This study was supported by funds from the Howard Hughes Medical Institute and the National Institutes of Health.
References
- 1.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nature Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 2.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 3.Davis L, Engebrecht J. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics. 1998;149:45–56. doi: 10.1093/genetics/149.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inagaki Y, Blouin C, Susko E, Roger AJ. Assessing functional divergence in EF-1alpha and its paralogs in eukaryotes and archaebacteria. Nucleic Acids Res. 2003;31:4227–4237. doi: 10.1093/nar/gkg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosoda N, et al. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem. 2003;278:38287–38291. doi: 10.1074/jbc.C300300200. [DOI] [PubMed] [Google Scholar]
- 6.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 7.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 8.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3′ -to-5′ degradation of mRNA. Mol Cell Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson JS, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 13.Inge-Vechtomov S, Zhouravleva G, Philippe M. Eukaryotic release factors (eRFs) history. Biol Cell. 2003;95:195–209. doi: 10.1016/s0248-4900(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 14.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 15.Carr-Schmid A, Pfund C, Craig EA, Kinzy TG. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol Cell Biol. 2002;22:2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong C, et al. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe. Mol Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- 17.van Hoof A. Conserved functions of yeast genes support the Duplication, Degeneration and Complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiba Y, et al. Evidence for autoregulation of cystathionine gamma-synthase mRNA stability in Arabidopsis. Science. 1999;286:1371–1374. doi: 10.1126/science.286.5443.1371. [DOI] [PubMed] [Google Scholar]
- 19.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 20.Onouchi H, et al. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005;19:1799–1810. doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem. 2004;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 22.Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 24.Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu Rev Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 25.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 28.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


