Abstract
In mice, zygotic or pro-B-cell-specific knock-out of the Pax5 gene allows differentiation of pro-B-cells into all hematopoietic lineages. We previously generated and characterized a murine B-cell lymphoma, dubbed Myc5, whose cells spontaneously lose Pax5 expression when cultured in vitro, but regain it when re-injected into syngeneic mice. In cultured Myc5 cells, the loss of Pax5 correlates with the acquisition of myeloid markers, such as CD11b and F4/80. Here, we sought to determine whether these cells are truly B-macrophage-restricted or, like Pax5-null progenitors, can give rise to additional hematopoietic lineages. In vitro differentiation assays with various cytokines showed that Myc5 cells do not differentiate into NK cells, dendritic cells, neutrophils, or osteoclasts. At the same time, in the presence of macrophage colony-stimulating factor (M-CSF), they readily phagocytose latex beads and provide T-cell help. Both phenomena are indicative of the bona fide macrophage phenotype. Conversely, enforced Pax5 re-expression in macrophage-like Myc5 cells led to down-regulation of the M-CSF receptor and re-acquisition of some B-cell surface markers (e.g., CD79a) and lineage-specific transcription factors (e.g., IRF4 and Blimp). Retrovirally encoded Pax5 also restored expression of several master B-cell differentiation proteins, such as the IL7 receptor and transcription factor E2A. In contrast, levels of EBF were unaffected by Pax5 suggesting that EBF acts exclusively upstream of Pax5 and might contribute to Pax5 expression. Indeed, transduction with an EBF-encoding retrovirus partly reactivated endogenous Pax5. Our data reveal the complex relationship between B-cell specific transcription factors and suggest the existence of numerous feedback mechanisms.
Introduction
Hematopoietic stem cells (HSC) can give rise to a multiplicity of lineages including B- and T-lymphoid cells, macrophages, natural killer cells, erythroid cells, dendritic cells, megakaryocytes, and osteoclasts. Development of these lineages requires the concerted functions of numerous transcription factors that control the differentiation of specific cell fates [2–6]. Uncommitted HSC and multipotential progenitors (MPP) often express low levels of mixed lineage patterns of gene expression [6,7]. Rare populations of cells are bipotential, able to give rise to two different lineages, such as the bipotential B-cell/macrophage progenitor observed in postnatal bone marrow [8]. Specific lineages may be dictated by induction of key cell fate determinants and the subsequent repression of alternative lineage-determining factors [6]. For instance, GATA-1 is crucial for erythroid development whereas PU.1 is needed for multiple hematopoietic lineages [9–12]. These factors can functionally antagonize each other’s activity [12,13]. Similarly, PU.1-to-C/EBPα ratios are crucial for regulating macrophage and neutrophil cell fates [14], and C/EBPα expression can drive B-cells toward the macrophage lineage [15].
In recent years, the relationship between the B- and macrophage lineages has become apparent. Initial suggestions of the close relationship between these lineages derived from studies with the 70Z/3 pre-B-cell line that can spontaneously differentiate into cells with a macrophage phenotype [16]. Other bipotential cells were subsequently characterized [17–20]. Expression level of transcription factor PU.1 was later found to be important for determining lineage commitment. High PU.1 levels in hematopoietic progenitors leads to macrophage differentiation, whereas low levels support B-cell development [21,22]. It has also been proposed that expression of Pax5 can support B-cell development by inhibiting the myeloid-specific functions of PU.1 [23,24], similar to the antagonistic relationship between PU.1 and GATA-1 during erythroid development [9,12]. Pax5 activates a number of genes important for the B-cell lineage while repressing alternative lineage genes, as well as those needed for plasma cell differentiation [25–29]. Pax5 is also crucial for rearrangement of distal immunoglobulin heavy chain variable genes, and knock-out of the Pax5 gene results in developmental arrest at the pro-B-cell stage [30–34]. Pax5 is unusual in that pro-B-cells from Pax5-null mice are plastic in their differentiated phenotype and can adopt numerous hematopoietic pathways [5,35].
We recently characterized a number of B-cell lymphomas derived by transduction of p53 null bone marrow cells with a c-Myc expressing retrovirus [36]. Injection of these transduced bone marrow cells into animal hosts resulted in development of B-cell lymphomas. Interestingly, some of these B-cell lymphomas had the ability to express macrophage markers when grown in vitro [37]. These lymphomas are typified by a line named Myc5 which showed the surprising ability to oscillate between the B-cell and macrophage lineages. When Myc5 cells are grown as a subcutaneous tumor in vivo, the cells retain a B-cell lymphoma phenotype. If grown in vitro on S17 stromal cells, Myc5 cells lose expression of surface markers characteristic of B-cells (CD45R and CD19) but gain expression of markers characteristic of macrophages (F4/80 and CD11b). Strikingly, Myc5 clones can repeatedly oscillate between these two lineages, thus exhibiting a surprising plasticity of differentiated stage [37]. This developmental plasticity is reminiscent of the behavior of previously identified bipotential cells [8], and of pax5 knock-out pro-B-cells which have the ability to adopt multiple hematopoietic lineages [5,35].
Since Pax5 expression is lost when Myc5 cells are grown in vitro [37], it was of interest to determine whether Myc5 cells have the capacity to differentiate into multiple hematopoietic lineages, or whether these cells are restricted to the bipotential B-cell-myeloid phenotype and yield bona fide macrophages capable of phagocytosis and T-cell help. Furthermore, this model afforded a unique opportunity to investigate the functional relationship between B-cell-specific transcription factors.
Methods and materials
Differentiation Assays
S17 bone marrow stromal cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. Feeder cells were irradiated with 1600 rad, and 2 hr later were plated with Myc5 cells harvested from tumors grown in C57BL6 mice. Media was supplemented with either 1 ng/ml IL-7 (B-cell conditions), 2 ng/ml M-CSF (macrophage conditions), 1 ng/ml GM-CSF (dendritic cell conditions), 5, 10, or 20 ng/ml TRANCE (osteoclast conditions), 1ng/ml IL-2 (NK cell conditions), or 0.2ng/ml IL-3, 0.1 ng/ml IL-6, 50ng/ml SCF for 3 wks followed by 6 days with 0.2 ng/ml G-CSF (neutrophil conditions) (all purchased from Peprotech). Cells were passaged on fresh irradiated S17 cells and fresh media and cytokines every 5 to 6 days for 26 days. Osteoclast differentiation was assessed by histochemical TRAP staining (Sigma) for tartrate-resistant acid phosphatase. Neutrophil differentiation was assessed by Giemsa staining followed by visual inspection. Alternatively, cells were cultured without a feeder layer, in the medium lacking IL-7 but supplemented with lipopolysaccharide (LPS, 10 μg/ml, Sigma-Aldrich, St. Louis, MO).
Fluorescent Activated Cell Sorting
Expression of cell surface markers was assessed at various time points by fluorescence activated cell sorting (FACS). Cells were washed and resuspended in PBS containing 0.1% BSA, then stained with various phycoerythrin-labeled antibodies (1 μg/ml per 106 cells) for 30 min on ice. Cells were washed and resuspended in FACS buffer and analyzed on a FACSCalibur flow cytometer. Data analyses were performed with Cellquest software. Antibodies used were: anti-mouse CD11b (Mac-1), anti-mouse NK1.1, anti-mouse CD45R (B220), anti-mouse CD40, or anti-mouse CD11c (all from Pharmingen.)
T cell Help assays
OVA specific CD4+ T cells were isolated from DO11.10 mice containing an MHC class II restricted TCR transgene specific for ovalbumen. Spleens were removed and were disrupted with sterile 25 gauge hypodermic needles in complete RPMI. Large debris was removed by filtration through sterile nylon mesh (70 μM pore diameter). The filtered cell suspension was first incubated with monoclonal antibodies (Pharmingen) against B220 (RA3-6B2), granulocytes (RB6-8C5), Class I MHC (M5/114), and CD8 (Ly-2, 53-6.7). Antibody-stained (non-CD4+) cells were removed by immunomagnetic depletion (Stem Cell Technologies, Vancouver, BC Canada). In order to determine the proliferative history of T cells in vivo, purified CD4+ lymphocytes were labeled with the fluorescent dye 5- 6-carboxyfluorescein diacetate succinimidyl ester (CFSE, Molecular Probes, Eugene, OR), which labels live cells and segregates equally between daughter cells during mitosis, as previously described [38]. CFSE labeled CD4+ lymphocytes were cultured with ovalbumen (1 μg/ml) and Myc5 cells differentiated under various conditions (5 T-cells: 1 Myc5 cell). CFSE fluorescence was analyzed by flow cytometry on day 4 of culture.
Phagocytosis assays
Phagocytosis assays were carried out as previously described [39] in 96-well plates in a total volume of 120 μl/well. Briefly, lymphoma cells (3.0 x 105 cells/well) were incubated with fluorescent beads (1.0-μm diameter YG, Polysciences) for 6 h in 5% CO2 at 37°C, at a cell/bead ratio of 1:10. After incubation and prior to collection, the cells were treated with trypsin-EDTA (0.05% Trypsin, Gibco) for 5 min at 37°C, then resuspended in PBS, pH 7.4. To remove adherent beads, cell suspensions were centrifuged (100 g, 10 min at 4°C) over a cushion of 3% BSA in PBS supplemented with 4.5% glucose. Cell pellets were resuspended in PBS (pH 7.4), and flow cytometric analyses were carried out using a fluorescence-activated cell analyzer (FACScan, Becton Dickinson). For each sample, 20,000 individual cells were analyzed and data were analyzed with Cell Quest™ software. Prior to flow cytometric analysis, a sample of the cell suspension was spun onto slides and stained with the Hema® 3 stain set according to the manufacture’s instructions (Fisher PROTOCOL).
Retroviral transduction
EBF and IL7R full-length cDNAs were expressed by the MigR1 retroviral vector. All retroviruses were generated using transfection with Lipofectamine 2000 (Invitrogen) into the GP293 packaging cells. Infections were carried out over the course of 36h in the presence of polybrene (4 μg/ml). Transduction efficiency was assessed using GFP fluorescence.
Quantitative RT-PCR
Total RNA was isolated from either 10 x 106 fresh Myc5 tumor cells, Myc5 cells differentiated into macrophages by culture on S17 feeder cells in media supplemented with M-CSF for 4–6 weeks, or Myc5 macrophage-like cells transduced with a Pax5-expressing retrovirus. Oligo(dT)-primed cDNA was made from 5 μg RNA using the Superscript cDNA synthesis kit (Strategene). Real-time PCR was performed in triplicate samples using primers specific for desired transcripts in the presence of SYBR Green. For each sample, cT’s corresponding to the gene of interest and actin were determined and adjusted by the same number of cycles, to make actin’s average cT equal 18. Assuming amplification efficiency of 100%, relative expression levels were expressed as 2[30-CT]. The nucleotide sequences of PCR primers were (5’-to-3’): CGGAAGCAGATGCGGGGAGAC (Pax5 sense), CTGTGACAATAGGGTAGGACTG (Pax5 antisense), CGGAAGCAGATGCGGGGAGAC (MCSFR sense), CTGTGACAATAGGGTAGGACTG (MCSFR anti-sense), CACGGTGAACTTGGGCGAGGAG (CD79a sense), GCTGTGATGATGCGGTTCTTGGTA (CD79a antisense), CGAGAAGCCGCAGACCAAACT (E2A sense), TCCCCGACCACGCCAGAC (E2A antisense), TGTCGGGACTTTGCGGAGAGG (Blimp1 sense), GGGTGAAATGTTGGAACGGTAGAT (Blimp1 antisense), GACTGCCGGCTGCATATCTGC (IRF4 sense), GCCGATCGCTGCACAGTGCCAG (IRF4 antisense), GCCAACAGCGAAAAGACC (EBF sense), GCTTGGAGTTATTGTGGAC (EBF anti-sense), AGCTGTTTCTGGAGAAAGTGG (IL7R sense), AACGACTTTCAGGTCAGAGGG (IL7R anti-sense), CACGGCATTGTCACTAACT (gamma-actin sense), CAGCCAGGTCCAGACGCAAGAT (gamma-actin antisense).
Immunoblotting
Pax5 levels were assessed in Myc5 cell lysates. Membranes were probed with a polyclonal anti-Pax5 antibody. The secondary antibody was used in a horseradish peroxidase-conjugated form (Amersham Biosciences, Piscataway, NJ). Antibody binding was detected using the enhanced chemiluminescence system (Amersham). A monoclonal antibody reactive with murine actin (Sigma-Aldrich) was used to confirm equal loading.
Results
Differentiation markers of Myc5 cells under various culture conditions
We had previously described the disappearance of B-cell markers from the surface of cultured Myc5 cells [37]. We now asked whether polyclonal B-cell activation might rescue the expression of these markers. To this end, cells were cultured either on the monolayer of S17 stromal cells or without the feeder layer but in the presence of lipopolysaccharide (LPS), a well-known polyclonal mitogen for both B- and T-cells [40]. As evidenced by data in Figure 1, the B-cell markers CD19 and B220 were concertedly lost regardless of the presence or the absence of LPS. At the same time, expression of the Mac1 antigen was maintained under both conditions.
Figure 1. Expression of lineage-specific cell surface markers on Myc5 cells following polyclonal stimulation.
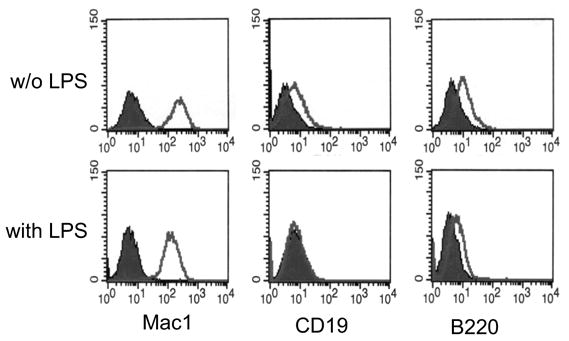
The top and bottom rows represent cells cultured without and with LPS, respectively. Individual panels represent stainings for Mac1, CD19, and B220 antigens, followed by flow cytometry. Filled and open plots represent unstained and antibody-stained cells, respectively.
To further test the differentiation capacity of Myc5 cells, Myc5 B-lymphoma cells were placed into various culture conditions known to enable pax5-null cells to remain B-cells or cross-differentiate into macrophages, dendritic cells, NK cells, neutrophils, or osteoclasts [5] (Figure 2). Culture aliquots were evaluated at several time points for cell lineage characteristics. Differentiation into various cell types was assessed by staining for cell surface markers, histochemical detection of lineage-specific proteins, or the analysis of cell morphology.
Figure 2. Schema for testing differentiation plasticity of Myc5 cells.
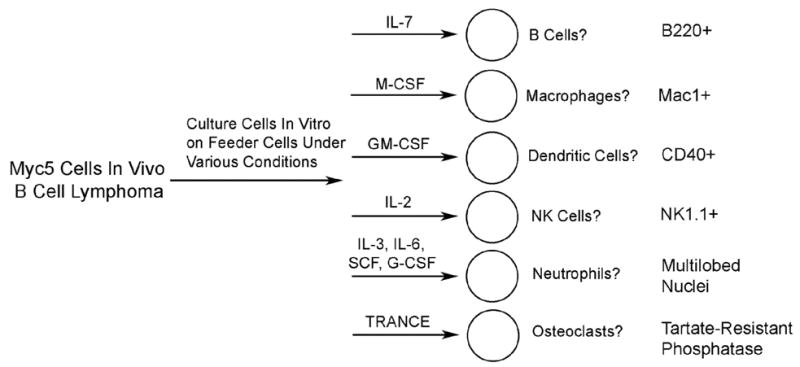
Shown are growth factors used, expected differentiation patterns, and lineage-specific markers.
First, Myc5 cells were cultured in the presence of IL-7, M-CSF, GM-CSF, or IL-2. These culture conditions had been reported to foster, respectively, the retention of B-cell phenotype and differentiation into macrophages, dendritic, or NK cells [5,35]. In all four conditions, Myc5 cells rapidly lost expression of the B-cell-specific surface marker B220 (Fig. 3A-D) and CD19 (not shown). It was particularly striking that Myc5 cells could not maintain B220 expression even in the presence of the B-cell mitogen IL-7. Instead, we observed Mac-1 expression characteristic of myeloid cells. While only 50% of cells were Mac-1-positive on day 16, essentially all cells were Mac-1 positive by day 26 (Figure 3A). When cultured in the presence of M-CSF, which promotes macrophage differentiation, most of cells were already Mac-1-positive on Day 16 (Figure 3B).
Figure 3. Expression of lineage-specific cell surface markers on Myc5 cells under different culture conditions.
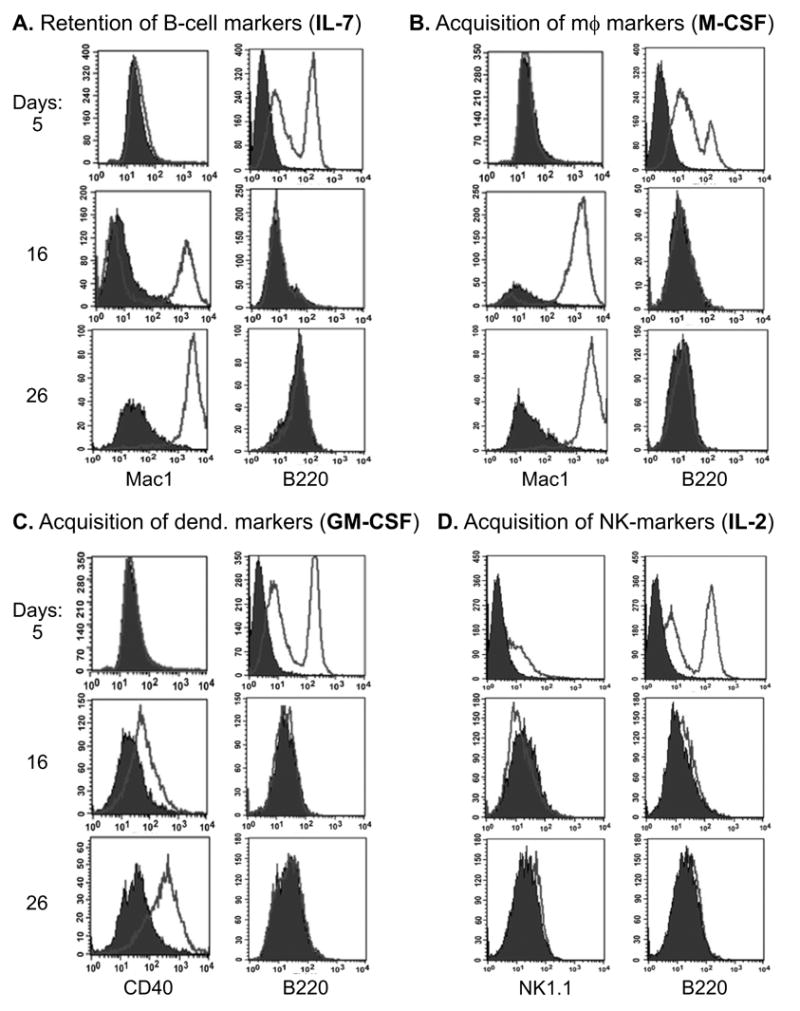
Panels A-D represent growth media supplemented with cytokines favoring particular lineages: B-cell (interleukin-7, or IL-7), macrophage (macrophage colony-stimulating factor, or M-CSF), dendritic (granulocyte-macrophage colony-stimulating factor, or GM-CSF), and natural killer (NK) cell (interleukin-7, or IL-2). B220, Mac-1, CD40, and NK1.1 are the corresponding surface markers. All cultures were assayed on days 5, 16, and 26 following cytokine stimulation using flow cytometry. Filled and open plots represent unstained and antibody-stained cells, respectively. For more details, see Methods and Materials.
We then assayed cells grown in GM-CSF and IL-2 for expression of CD40 and NK1.1 markers, respectively. While very little CD40 expression was observed in IL-7 treated cells (data not shown), GM-CSF treatment resulted in most cells being positive although the levels of CD40 were relatively low, especially on day 16 (Figure 3C). When cultured in IL-2, NK1.1 expression was not induced even after 26 days, indicating the absence of NK cell phenotype (Figure 3D). Thus, it appears that Myc5 cells have an inherent propensity to adopt a macrophage-like phenotype, with some expression of the dendritic marker CD40, but only when grown in the presence of GM-CSF.
Additionally, Myc5 cells were cultured in the presence of either TRANCE or the cytokine cocktail consisting of IL-3, IL-6, SCF, and G-CSF. These conditions are known to promote osteoclast and neutrophil differentiation in Pax5-null cells. However, TRANCE-treated Myc5 cells failed to express tartate-resistant phosphatase characteristic of osteoclasts (data not shown). Similarly, IL-3/IL-6/SCF/G-CSF treatment did not result in the emergence of cells with multi-lobated nuclei characteristic of neutrophils (data not shown). Thus, Myc5 cells appear restricted to the B-macrophage phenotype. To corroborate this conclusion, we tested their ability to provide T-cell help and phagocytose latex beads.
T-cell help by Myc5 cells
To determine whether Myc5 cells can provide T-cell help, they were pre-treated with various growth factors, pulsed with the ovalbumin peptide (OVA), and incubated with OVA-specific CD4+ T cells isolated from spleens of D011.10 OVA-transgenic mice. T-cell proliferation was monitored using as a tracer intracellular dye CFSE as described in Materials and Methods. We found that under no conditions was proliferation observed in the absence of the cognate antigen (Figure 4, “w/o OVA” column). In the absence of any phenotypic changes (IL2- and IL-3/6, G-/CSF-pretreated cells from Figure 3), Myc5 cells provided weak but detectable T-cell help: most T-cells were proliferating but the most populous fraction was still M1 (undivided cells) (Figure 4A and C).
Figure 4. T cell help provided by Myc5 cells under different culture conditions.
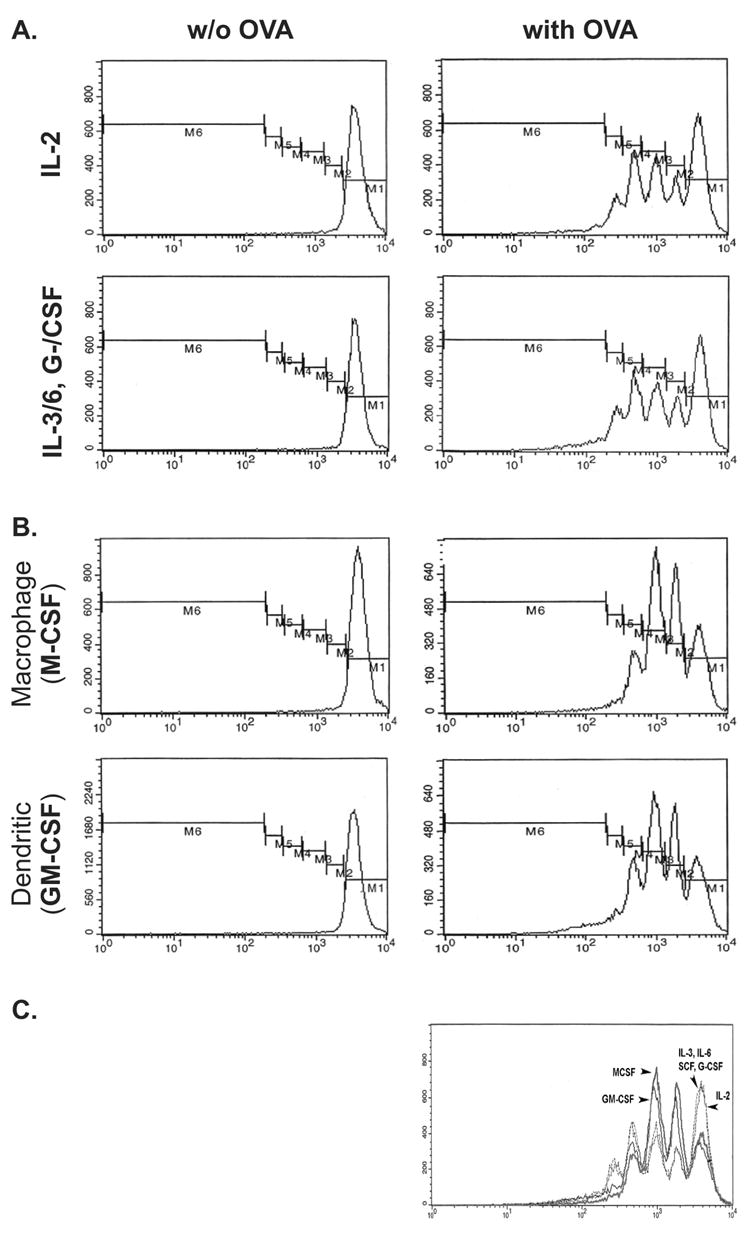
Myc5 cells were grown under conditions favoring NK- (IL-2) or neutrophil (IL-3/6, G-/CSF) (A) and macrophage (M-CSF) or dendritic (GM-CSF) (B) differentiation. Cells were assayed for their ability to provide T cell help by measuring proliferation of OVA-specific T cells in the absence (left panels) or presence (right panels) of OVA-primed cells. Profiles show proliferation of T-cells, as evidenced by dilution of the intracellular dye CFSE (from M1 to M6). The C panel shows composite traces of T-cell proliferation under all 4 conditions. Arrows point at fractions with the most cells.
M-CSF and GM-CSF pretreatments, which boost macrophage and macrophage +dendritic phenotype respectively, significantly increased T cell help: the most populous fraction was now M3 (twice divided cells) (Figure 4B and C). Interestingly, although dendritic cells are better antigen-presenting cells than macrophages, GM-CSF-pretreated Myc5 cells provided the same degree of help as M-CSF-pretreated. This finding suggests that Myc5 cells can be forced to differentiate primarily into macrophages (either by M-SCF or GM-CSF) but not into dendritic cells.
Phagocytic ability
Another key functional property of macrophages is their ability to phagocytose foreign particles, such as latex beads. Therefore, we compared phagocytosis ability of Myc5 cells to that of similarly derived Myc3 lymphoblasts which maintain their B-cell identity in vitro [36,37]. Specifically, we incubated M-CSF-differentiated Myc5 and Myc3 cells with fluorescent latex beads. After an hour, cells were subjected to FACS analysis to detect cell-associated fluorescence due to ingested beads. As expected, M-CSF differentiated Myc5 cells were labeled much more frequently (54% vs. 0.8%) and with greater intensity than Myc3 cells (Figure 5, top panels). Furthermore, phase-contrast microscopy of the same samples indicated that the beads were truly internalized in Myc5 macrophages, but merely sticking to the cell surface in occasional fluorescently labeled Myc3 cells (Figure 5, bottom panels). Thus, M-CSF-stimulated Myc5 cells can function as bona fide phagocytic cells.
Figure 5. Phagocytosis by Myc5 cells.
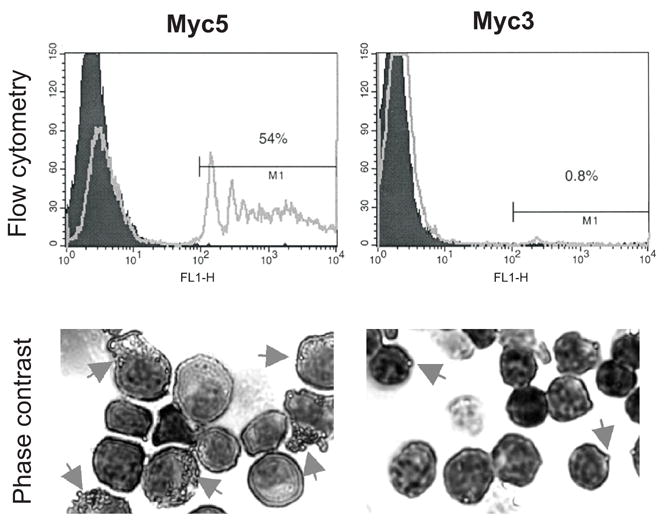
Myc5 and control Myc3 cells were grown in media supplemented with M-CSF. Cells pre-incubated with fluorescent latex beads were subjected to flow cytometry (top panels) and phase-contrast microscopy (bottom panels). In the top panels, filled and open plots refer to cells without and with beads, respectively. In the bottom panels, arrows point at cells with ingested or attached beads. For more details, see Materials and Methods.
Reversion of lineage-specific gene transcription upon restoration of Pax5 expression
To determine whether the absence of Pax5 is required for the macrophage phenotype, we transduced cultured Myc5 cells with a Pax5-encoding MIGR1 retrovirus [37]. In these cells, levels of Pax5 mRNA were several-fold higher than in Myc5 tumors, but no gross overexpression was attained (Figure 6, “Pax5” panel). Vector-transduced cells expressed no detectable Pax5. Consistent with this expression pattern, control Myc5+GFP cells maintained M-CSFR expression, but this macrophage-specific gene was down-regulated in Myc5+Pax5/GFP cells (“M-CSFR” panel). Conversely, B-cell specific CD79a gene was down-regulated in Myc5+GFP cells but its expression was fully restored by retrovirally encoded Pax5 (“CD79a” panel). Similar expression patterns were observed for genes encoding B-cell transcription factors IRF4 and Blimp, although Blimp mRNA levels were not fully restored by retrovirally encoded Pax5 (compare “IRF4” and “Blimp” panels). Interestingly, Blimp and Pax5 are known to cross-repress each other in plasma cells ([29,41] and Figure 6B).
Figure 6. Lymphoid- and macrophage- specific gene expression is affected by Pax5.
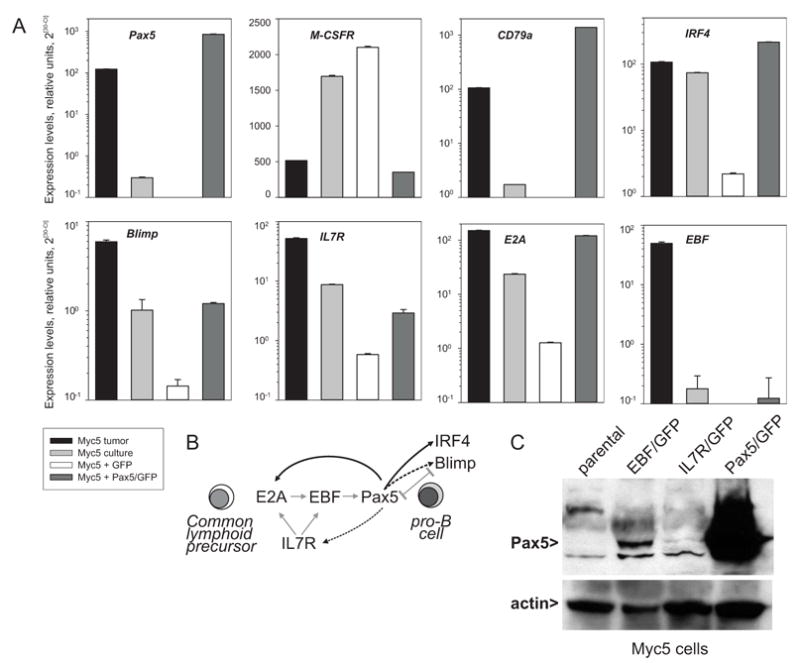
A Real-time RT-PCR analysis of Pax5, M-CSFR, CD79a, IRF4, Blimp, IL7R, E2A, and EBF transcript levels in the Myc5 tumor, cultured Myc5 cells, and cultured Myc5 cells transduced with the empty MIGR1 vector (“GFP”) or the Pax5/MIGR1 virus (“Pax5/GFP”). Gamma-actin was used as an internal control. Relative expression levels were calculated as described in Methods and Materials. B. Interactions between IL7R and B-lymphoid transcription factors during differentiation of common lymphoid progenitor (CLP) towards the pro-B-cell. Grey arrows indicate known interactions (modified from [1]). Black arrows refer to data in A. Dotted arrows indicate partial restoration of mRNA levels by retrovirally encoded Pax5. C. Immunoblotting with anti-Pax5 and anti-actin antibodies. Cells used were parental Myc5 cells and their EBF-, IL7R-, and Pax5-transduced derivatives.
We also asked whether Pax5 affects expression of several master B-cell differentiation proteins, such as the interleukin-7 receptor (IL7R) and transcription factors E2A and EBF. All three are presumed to act upstream of Pax5 during differentiation of common lymphoid progenitor towards pro-B-cell ([1,42] and Figure 6B). Nevertheless, levels of both E2A and IL7R transcripts were significantly elevated following Pax5 transduction, although levels of IL7R mRNA have not recovered completely (compare “E2A” and “IL7R” panels). In contrast, levels of EBF were unaffected by Pax5 and remained as low in Pax5-transduced as in parental Myc5 cells (“EBF” panel). This suggested that only EBF is a bona fide upstream regulator of Pax5 and that its loss might contribute to Pax5 silencing.
To address this possibility, we transduced cultured Myc5 cells with retroviruses encoding murine EBF and IL7R. Acutely infected cultures were tested for Pax5 re-expression using immunoblotting. As anticipated, Pax5 protein expression was detectable, albeit weakly, in EBF-, but not IL7R-transduced cells (Figure 6C). Our data are in agreement with those obtained with EBF-transduced 70Z/3 pre-B lymphocytes [43].
Discussion
Myc5 cells contain rearranged Ig genes and in vivo express genes characteristic of the B-cell lineage (B220, Igκ, Pax5, IRF4, CD79a). On the other hand, when grown in vitro, expression of these genes is drastically decreased and myeloid-specific genes are reactivated [34,44]. Myc5 cells grown in vitro also develop the ability to provide T cell help. Identical T-cell help capacity was observed by Myc5 cells grown in either GM-CSF or M-CSF supporting the conclusion that in vitro grown Myc5 cells are primarily macrophages rather than dendritic cells. In addition, Myc5 cells grown in M-CSF developed the ability to efficiently phagocytose latex beads. Thus, their cell surface markers, RNA transcripts, T-cell help and phagocytosis abilities all indicate that in vitro Myc5 cells readily adopt the macrophage phenotype. Interestingly, PU.1 expression levels are quite similar in B- and macrophage-like Myc5 cells (data not shown), at variance with previous reports indicating that macrophages generally express elevated PU.1 levels compared to B-cells [21,22]. Also unchanged are C/EBPα levels (data not shown), despite its prominent role in determining B-lymphoid vs. myeloid cell fate [15]
The acquisition of the macrophage phenotype is preceded by the loss of Pax5 expression [37]. However, several features make Myc5 cells rather unique among other Pax5 non-expressors of B-cell origin. Most notably, our data reveal their relatively limited differentiation capacity. Unlike Pax5-null cells [5], Myc5 cells in culture were unable to adopt numerous hematopoietic lineages and become NK cells, osteoclasts, or neutrophils under the conditions tested. Nor did loss of Pax5 result in an increase in plasma cell-specific transcripts such as Blimp-1 (unpublished observations), as was recently observed in Pax5-deficient DT40 cells [29]. Thus, although Myc5 cells lose Pax5 expression when adopting the macrophage lineage, these cells are not as multipotent as Pax5-null cells. Rather, they remain committed to the B-macrophage bifurcation. In addition, unlike Pax5-null pro-B-cells, reintroduction of retroviral expressed Pax5 does not completely rescue the B-cell phenotype, as evidenced by the lack of CD19 expression [37]. This is despite the fact that at least two linage-specific transcription factors, IRF4 and E2A, were fully reactivated, and the levels of IL7R were significantly elevated (Figure 6). The differentiation status of these cells warrants further investigation.
A number of cell lines or bone marrow cells have shown the ability to give rise to B-cells and macrophages [16–20]. A splenic B-cell population grown on fibroblasts simultaneously expresses B-cell and macrophage characteristics, and a rare human bone marrow population is bipotential, capable of generating both B-cells and macrophages [8,45]. The later population is similar to Myc5 cells in that they cannot generate NK or dendritic cells. However, these cells, unlike Myc5 lymphoblasts, lack VDJH rearrangements [8]. Thus, Myc5 may represent a more mature cell derived from this bipotential lineage, which still retains trans-differentiation capacity, even after immunoglobulin gene rearrangement. Alternatively, Myc5 cells may represent a distinct lineage of hematopoietic cells that retains both B-cell and macrophage properties even after terminal differentiation. In any event, repeated isolation of hematopoietic tumors with lineage infidelity following Myc-induced transformation of bone marrow cells ([37] and data not shown) argues that this is a significant mammalian hematopoietic lineage with potential to oscillate between two developmental fates depending upon environmental cues. Notably, a human B-cell lymphoma has been described that responded to therapy, only to relapse as a histiocytic sarcoma with rearranged immunoglobulin genes [46]. Thus, studies on Pax5-controlled cell differentiation can offer important insights into mechanisms of tumorigenesis as well.
Acknowledgments
We thank Drs Rodney DeKoter (University of Cincinnati) and James Hagman (National Jewish Medical & Research Center) for their gifts of the IL7R- and EBF-encoding retroviruses, respectively. This work was supported by NIH grants GM42415 (M.L.A) and CA102709 (A.T.-T.) and the Commonwealth of Pennsylvania Health Research Formula Fund #4100020574.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Roessler S, Grosschedl R. Role of transcription factors in commitment and differentiation of early B lymphoid cells. Semin Immunol. 2006;18:12–19. doi: 10.1016/j.smim.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Glimcher LH, Singh H. Transcription factors in lymphocyte development - T and B cells get together. Cell. 1999;96:13–23. doi: 10.1016/s0092-8674(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 3.Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11:513–519. doi: 10.1016/s0959-437x(00)00226-4. [DOI] [PubMed] [Google Scholar]
- 4.Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 2000;14:2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 6.Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci USA. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 8.Montecino-Rodriguez E, Leathers H, Dorshkind K. Bipotential B-macrophage progenitors are present in adult bone marrow. NatImmun. 2001;2:83–88. doi: 10.1038/83210. [DOI] [PubMed] [Google Scholar]
- 9.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 11.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- 14.Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 15.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 16.Paige CJ, Kincade PW, Ralph P. Murine B cell leukemia line with inducible surface immunoglobulin expression. J Immunol. 1978;121:641–647. [PubMed] [Google Scholar]
- 17.Principato M, Cleveland JL, Rapp UR, Holmes KL, Pierce JH, Morse HC, Klinken SP. Transformation of murine bone-marrow cells with combined v-Raf-v-Myc oncogenes yields clonally related mature B-cells and macrophages. MolCellBiol. 1990;10:3562–3568. doi: 10.1128/mcb.10.7.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson WF, Pierce JH, Holmes KL. Evidence for a developmental relationship between CD5+ B-lineage cells and macrophages. Annals NY Acad Sci. 1992;651:112–129. doi: 10.1111/j.1749-6632.1992.tb24601.x. [DOI] [PubMed] [Google Scholar]
- 19.Holmes KL, Pierce JH, Davidson WF, Morse HC., III Murine hematopoietic cells with pre-B or pre-B/myeloid characteristics are generated by in vitro transformation with retroviruses containing fes, ras, abl, and src oncogenes. J Exp Med. 1986;164:443–457. doi: 10.1084/jem.164.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M, Strasser A, Baumgarth N, Cicuttini FM, Welch K, Salvaris E, Boyd AW. A novel cellular model (SPGM 1) of switching between the pre-B cell and myelomonocytic lineages. J Immunol. 1993;150:4395–4406. [PubMed] [Google Scholar]
- 21.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 22.Nutt SL, Metcalf D, D'Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–231. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maitra S, Atchison M. BSAP can repress enhancer activity by targeting PU.1 function. Mol Cell Biol. 2000;20:1911–1922. doi: 10.1128/mcb.20.6.1911-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang MY, Monroe JG. BSAP/Pax5A expression blocks survival and expansion of early myeloid cells implicating its involvement in maintaining commitment to the B-lymphocyte lineage. Blood. 1999;94:3621–3632. [PubMed] [Google Scholar]
- 25.Fitzsimmons D, Hodsdon W, Wheat W, Maira SM, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 26.Nutt SL, Morrison AM, Dorfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 1998;17:2319–2333. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol Cell Biol. 1992;12:2662–2672. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24:269–281. doi: 10.1016/j.immuni.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Nera KP, Kohonen P, Narvi E, Peippo A, Mustonen L, Terho P, Koskela K, Buerstedde JM, Lassila O. Loss of Pax5 promotes plasma cell differentiation. Immunity. 2006;24:283–293. doi: 10.1016/j.immuni.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5' IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nutt SL, Thevenin C, Busslinger M. Essential functions of Pax-5 (BSAP) in pro-B cell development. Immunobiol. 1997;198:227–235. doi: 10.1016/S0171-2985(97)80043-5. [DOI] [PubMed] [Google Scholar]
- 33.Hsu LY, Liang HE, Johnson K, Kang C, Schlissel MS. Pax5 activates immunoglobulin heavy chain V to DJ rearrangement in transgenic thymocytes. J Exp Med. 2004;199:825–830. doi: 10.1084/jem.20032249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson K, Pflugh DL, Yu D, Hesslein DG, Lin KI, Bothwell AL, Thomas-Tikhonenko A, Schatz DG, Calame K. B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5. Nat Immunol. 2004;5:853–861. doi: 10.1038/ni1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–113. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- 36.Yu D, Thomas-Tikhonenko A. A non-transgenic mouse model for B-cell lymphoma: in vivo infection of p53-null bone marrow progenitors by a Myc retrovirus is sufficient for tumorigenesis. Oncogene. 2002;21:1922–1927. doi: 10.1038/sj.onc.1205244. [DOI] [PubMed] [Google Scholar]
- 37.Yu D, Allman D, Goldscmidt M, Atchison M, Monroe JG, Thomas-Tikhonenko A. Oscillation between B-lymphoid and myeloid lineages in Myc-induced hematopoietic tumors following spontaneous silencing/reactivation of the EBF/Pax5 pathway. Blood. 2003;101:1950–1955. doi: 10.1182/blood-2002-06-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells AD, Gudmundsdottir H, Turka LA. Following the fate of individual T cells throughout activation and clonal expansion. Signals from T cell receptor and CD28 differentially regulate the induction and duration of a proliferative response. J Clin Invest. 1997;100:3173–3183. doi: 10.1172/JCI119873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Peters R, Lapatra S, Vazzana M, Sunyer JO. Anaphylatoxin-like molecules generated during complement activation induce a dramatic enhancement of particle uptake in rainbow trout phagocytes. Dev Comp Immunol. 2004;28:1005–1021. doi: 10.1016/j.dci.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Greaves M, Janossy G, Doenhoff M. Selective triggering of human T and B lymphocytes in vitro by polyclonal mitogens. J Exp Med. 1974;140:1–18. doi: 10.1084/jem.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busslinger M. Transcriptional control of early B cell development. AnnuRev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 43.Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodawadekar S, Wei F, Yu D, Thomas-Tikhonenko A, Atchison M. Epigenetic histone modifications do not control Igk locus contraction and intranuclear localization in cells with dual B cell-macrophage potential. J Immunol. 2006 doi: 10.4049/jimmunol.177.9.6165. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borrello MA, Phipps RP. The B/macrophage cell: An elusive link between CD5(+) B lymphocytes and macrophages. Immunol Today. 1996;17:471–475. doi: 10.1016/0167-5699(96)20031-b. [DOI] [PubMed] [Google Scholar]
- 46.Feldman AL, Minniti C, Santi M, Downing JR, Raffeld M, Jaffe ES. Histiocytic sarcoma after acute lymphoblastic leukaemia: a common clonal origin. Lancet Oncol. 2004;5:248–250. doi: 10.1016/S1470-2045(04)01428-7. [DOI] [PubMed] [Google Scholar]


