Abstract
Transposable elements are a major mutation source and powerful agents of adaptive change. Some transposable element insertions in genomes increase to a high frequency because of the selective advantage the mutant phenotype provides. Cyp6g1-mediated insecticide resistance in Drosophila melanogaster is due to the upregulation of the cytochrome P450 gene Cyp6g1, leading to the resistance to a variety of insecticide classes. The upregulation of Cyp6g1 is correlated with the presence of the long terminal repeat (LTR) of an Accord retrotransposon inserted 291bp upstream of the Cyp6g1 transcription start site. This resistant allele (DDT-R) is currently at a high frequency in D. melanogaster populations around the world. Here, we characterize the spatial expression of Cyp6g1 in insecticide-resistant and -susceptible strains. We show that the Accord LTR insertion is indeed the resistance-associated mutation and demonstrate that the Accord LTR carries regulatory sequences that increase the expression of Cyp6g1 in tissues important for detoxification, the midgut, Malpighian tubules, and the fat body. This study provides a significant example of how changes in tissue-specific gene expression caused by transposable-element insertions can contribute to adaptation.
TRANSPOSABLE elements (TEs), present in most sequenced genomes to date, were once referred to as “selfish DNA,” alluding to the presumptively parasitic nature of these highly repetitive elements (Doolittle and Sapienza 1980; Orgel and Crick 1980). There is now increasing evidence that TEs play an important role in driving and shaping genome evolution (Kazazian 2004). For example, in humans it has been shown that TEs contribute a substantial number of regulatory sequences (Britten 1996; Jordan et al. 2003) and form part of the coding sequences of some genes (Britten 2004). There are numerous types of mutations associated with TE insertions. TEs can cause mutations by inserting into the coding regions of genes and disrupting or altering gene function. There are also examples of TEs inserting into the untranslated regions (UTRs) of genes, changing transcript length and stability (Landry et al. 2002; Dunn et al. 2003; Marsano et al. 2005). Insertions of TEs into the regulatory regions of genes can also disrupt or alter gene expression, by introducing regulatory elements such as enhancers (Argeson et al. 1996; Conte et al. 2002) or insulators (Gdula et al. 1996; Conte et al. 2002; Parnell et al. 2003).
Insecticide resistance is a model for studying evolutionary change, as the selective agent is known (the insecticide), and the response to selection (the evolution of resistance) is usually rapid (McKenzie and Batterham 1994). There has been a long predicted role of TEs in the evolution of insecticide resistance (Wilson 1993). However, it is only recently that resistance mechanisms potentially involving TEs have been identified in natural insect populations (ffrench-Constant et al. 2006), where resistance is associated with either the deletion or the inactivation of an insecticide target protein (Gahan et al. 2001), the truncation of a gene resulting in the generation of a protein with novel function (Aminetzach et al. 2005), or the increased expression of a gene associated with metabolic insecticide resistance (Daborn et al. 2002; Schlenke and Begun 2004; Marsano et al. 2005). Both for insecticide resistance examples and for other examples of TE-associated insertions, rarely has the effect of the mutation caused by the TE been fully characterized at the molecular level.
In Drosophila melanogaster, insecticide resistance mapping to the DDT-R locus is due to the overexpression of the cytochrome P450 gene Cyp6g1 (Daborn et al. 2001). In populations collected from different locations around the world, increased expression of Cyp6g1 is correlated with the presence of the 491-bp long terminal repeat (LTR) of an Accord retrotransposon 291 bp upstream from the transcription start site of this gene (Daborn et al. 2002; Catania et al. 2004). In this study we investigate if the insertion of the Accord LTR is responsible for the increased expression of Cyp6g1. We characterize the expression pattern of Cyp6g1 in third instar larvae of strains with and without the Accord LTR insertion upstream of Cyp6g1 and demonstrate that the molecular mechanism of Cyp6g1 overexpression is due to the Accord LTR insertion. We also demonstrate that the Accord LTR carries regulatory sequences that change the spatial expression of Cyp6g1, resulting in an insecticide resistance phenotype.
MATERIALS AND METHODS
Drosophila strains and insecticide bioassays:
All Drosophila strains were maintained on standard medium at 25°. Unless otherwise indicated, all strains were obtained from the Bloomington Drosophila Stock Center (Indiana University). DNA constructs were transformed into the w1118 strain using standard techniques (Rubin and Spradling 1982; Spradling and Rubin 1982). The w1118 strain does not contain the Accord LTR insertion upstream of Cyp6g1 (data not shown). Expression of Cyp6g1 in transgenic flies was achieved using the GAL4-UAS system (Brand and Perrimon 1993). UAS-Cyp6g1 strains (Daborn et al. 2002) were generated by cloning the open reading frame of Cyp6g1 from the Canton-S strain into the pUAST vector. Independent transformants in a w1118 background were then isolated using standard techniques. The 6g1HR-GAL4-6c strain (generated and characterized in this study) carries 1608 bp to the translational start of Cyp6g1 from the Hikone-R strain cloned upstream of GAL4 and inserted onto chromosome III. This strain expresses GAL4 in the gastric cecum, midgut, Malpighian tubules, and fat body of larvae and was used to drive Cyp6g1 in these tissues in crosses to UAS-Cyp6g1 strains. Insecticide resistance bioassays using dichloro–diphenyl–trichloroethane (DDT), dicyclanil, and nitenpyram were performed using standard techniques (Daborn et al. 2001; Pyke et al. 2004; Magoc et al. 2005). Survival to adulthood from five replicates of 50 larvae (first instar) per vial was used to assay for resistance to nitenpyram and dicyclanil. For DDT, five replicates of 20 adult females (4 days posteclosion) were used in a 24-hr contact assay (Daborn et al. 2001).
In situ hybridization:
The open reading frame (ORF) of Cyp6g1 was amplified from y; cn bw sp cDNA by PCR using Taq polymerase (Promega, Madison, WI) and the primers ORF6g1F (CGA CAG CGG CCG CAT GGT GTT GAC CAG GT) and ORF6g1R (GCG ATT CTA GAT CAT TGG AGC GAT GGA CC). The resulting PCR product was cloned into pGEM-T Easy (Promega) in both sense and antisense orientations with respect to the T7 RNA polymerase transcription initiation site. Plasmid constructs were then linearized with SalI and digoxigenin labeled using Megascript T7 polymerase (Ambion) and DIG-labeled dNTP mix (Roche, Indianapolis) following the manufacturer's instructions. The final concentration and purity of probes was determined by UV spectrophotometry and agarose gel electrophoresis. Third instar larvae were dissected in PBS and fixed in 8% paraformaldehyde. In situ hybridization was performed as previously described (Tautz and Pfeifle 1989).
Real-time PCR:
Malpighian tubules, midgut, and fat bodies were hand dissected from 200 feeding and wandering third instar larvae. RNA isolation and real-time PCR were performed as previously described (Bogwitz et al. 2005). PCR primers used were RpL11F (CGA TCC CTC CAT CGG TAT CT) and RpL11R (AAC CAC TTC ATG GCA TCC TC) for RpL11 and Cyp6g1aF (GCC CGC TGC GAT CCC CAT) and Cyp6g1aR (CCT TTC CAA TCT CCT GCA TA) for Cyp6g1.
Reporter constructs:
Transgenic strains carrying Cyp6g1 promoter regions in front of GAL4 were generated from different strains to determine if differences in Cyp6g1 expression between strains were the result of differences in Cyp6g1 promoter DNA sequence. DNA from insecticide-resistant Accord LTR carrying strain Hikone-R and two different insecticide-susceptible strains not carrying the Accord LTR insertion (Canton-S and y; cn bw sp) were used in this study. The upstream region of Cyp6g1 consisting of 1608 bp to the translational start site was PCR amplified from Hikone-R using the Expand High-Fidelity PCR system (Roche) and the primers MBF1 (ATT TGA TCC CGT CAT TTC GCC) and 5p2R (TTT GGG GAT GTC GAT GTA ATG). The equivalent 1200- and 1197-bp fragments were amplified from Canton-S and y; cn bw sp, respectively, using the same primers. The Hikone-R and Canton-S fragments were cloned into the plasmid pGEM-T Easy (Promega) and subcloned into pBC SK− (Stratagene, La Jolla, CA), using the EcoRI restriction sites. The GAL4 gene from pGATB (gift from Lucy Cherbus, Indiana University) was ligated into pBC SK− containing the promoter fragment via the BamHI and NotI restriction sites. The promoter fragment and the GAL4 gene were then ligated into the Drosophila transformation vector pW8, using the KpnI and NotI restriction sites. The 1197-bp upstream region of Cyp6g1 from y; cn bw sp was also cloned directly in front of GFP.nls in the pStinger vector, using the EcoRI restriction site (Barolo et al. 2000).
To determine if the Accord LTR by itself could drive Cyp6g1 expression, the 491-bp Accord LTR was PCR amplified from the Hikone-R strain, using primers accordF (CGT GAG TTA CGG GTG CCT CCG) and accordR (AGT TAC CAT GCC CAG CAT TAA C). This fragment was cloned into the pGEM-T Easy vector (Promega), sequenced, and subcloned in both directions into pH-Stinger, which contains a minimal Hsp70 promoter, and pStinger, which does not contain a minimal Hsp70 promoter (Barolo et al. 2000). Final constructs were microinjected (0.5 μg/μl) into <1-hr-old embryos of the w1118 strain, along with a Δ2–3 transposase source (0.1 μg/μl) as per standard procedures (Rubin and Spradling 1982; Spradling and Rubin 1982). The w1118 strain does not contain the Accord LTR insertion upstream of Cyp6g1 (data not shown). Independent transformed lines were made homozygous and the inserted construct was mapped to a chromosome using the w; If/CyO; MKRS/TM6b,Tb strain (gift from G. Hime, University of Melbourne). Expression in at least three independent transformants was compared. All GFP images were captured using the SZX12 stereomicroscope system (Olympus, Lake Success, NY).
5′ RACE:
To determine the start site of Cyp6g1 transcription in strains carrying the Accord LTR and wild-type strains, 5′ RACE was conducted using a SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA) and a primer within exon 1 of Cyp6g1 (CAG TGC GGC GAC CAC CAA AAG AG).
RESULTS
Increased expression of Cyp6g1 is tissue specific:
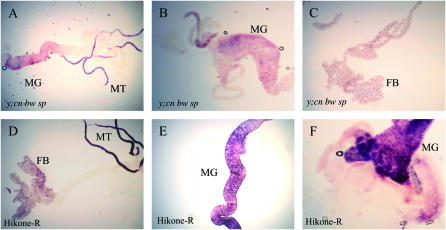
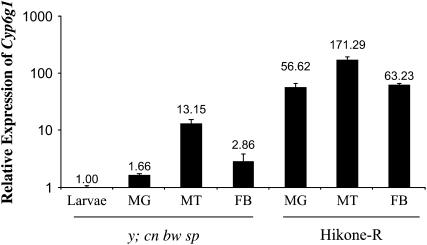
To determine the spatial distribution of increased expression of Cyp6g1 in insecticide-resistant flies, in situ hybridization of a DIG-labeled antisense Cyp6g1 probe was conducted in third instar larvae. In the insecticide-susceptible strain y; cn bw sp, Cyp6g1 mRNA was detected in parts of the midgut and in the Malpighian tubules in early third instar larvae. In wandering third instar larvae, Cyp6g1 mRNA was detected in parts of the midgut, the Malpighian tubules, and the fat body (Figure 1, A–C). In the insecticide-resistant strain Hikone-R that carries the Accord LTR inserted upstream of Cyp6g1 (Daborn et al. 2002), Cyp6g1 mRNA was detected in the gastric cecum, the entire midgut, the Malpighian tubules, and the fat body in both feeding and wandering third instar larvae (Figure 1, D–F). Additionally, the intensity of staining in these three tissues is higher in Hikone-R when compared to y; cn bw sp. Cyp6g1 mRNA was not detected in the hindgut, salivary glands, central nervous system, or any of the imaginal discs in either of these strains. To quantify the abundance of Cyp6g1 mRNA in larvae with and without the Accord LTR insertion, real-time PCR was performed on RNA isolated from midguts, Malpighian tubules, and fat body dissected from both feeding and wandering third instar larvae. Increases in the abundance of Cyp6g1 mRNA in the midgut, Malpighian tubules, and the fat body were detected in Hikone-R, when compared to y; cn bw sp at a magnitude of ∼10–40 folds in each tissue measured (Figure 2).
Figure 1.—
In situ hybridization of Cyp6g1 antisense digoxgenin-labeled RNA probe on tissues dissected from y; cn bw sp and Hikone-R third instar wandering larvae. (A–C) Third instar larvae of y; cn bw sp. Cyp6g1 mRNA is detected in midgut (MG), Malpighian tubules (MT), and fat body (FB). (D–F) Third instar larvae of Hikone-R. Cyp6g1 mRNA is detected in midgut, Malpighian tubules, and fat body. Staining is more intense in Hikone-R than in y; cn bw sp, suggesting that Cyp6g1 mRNA is present in these tissues at a higher level.
Figure 2.—
Real-time PCR on dissected tissues from third instar larvae. All values are relative to RPL11 and normalized to y; cn bw sp whole third instar larvae. An increase in Cyp6g1 mRNA is detected in the midgut (MG), Malpighian tubules (MT), and the fat body (FB) of Hikone-R. Values are the mean of two replicates ± SEM.
Characterization of Cyp6g1 expression using transgenes:
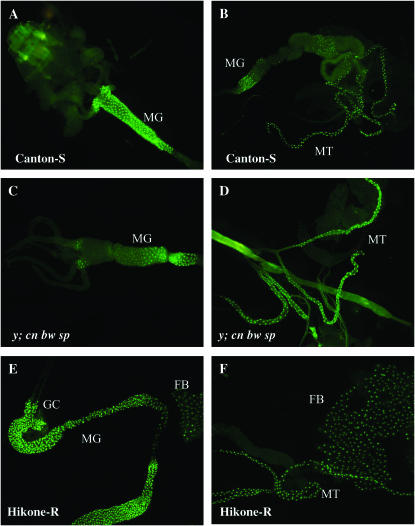
To determine if the 5′ upstream region of Cyp6g1 contains all the essential elements for the expression of Cyp6g1 in third instar larvae, transgenic reporter constructs containing the upstream regions of Cyp6g1 were constructed. In strains driving GFP under the control of the Cyp6g1 promoter of Canton-S, GFP was detected in sections of the midgut and the Malpighian tubules in both feeding and wandering third instar larvae (Figure 3, A and B). In strains driving GFP under the control of the Cyp6g1 promoter of y; cn bw sp, GFP was detected in parts of the midgut, in the Malpighian tubules in feeding third instar larvae, and additionally in the fat body in late third instar larvae (Figure 3, C and D). Finally, strains driving GFP under the control of the Cyp6g1 promoter of Hikone-R express GFP in the gastric cecum, the entire midgut, Malpighian tubules, and the fat body in both feeding and wandering third instar larvae (Figure 3, E and F). The intensity of GFP expression was much higher in the Hikone-R-driven strains than in the other two strains. The upstream region of Cyp6g1 is sufficient to replicate the native expression of Cyp6g1 in third instar larvae as detected using in situ hybridization in both Accord (Hikone-R) and non-Accord (y; cn bw sp) strains and quantified using real-time PCR. This indicates that all of the elements required for Cyp6g1 expression in third instar larvae are located within 1.2 kb upstream of the translational start.
Figure 3.—
Promoter constructs of Cyp6g1 from Canton-S, y; cn bw sp, and Hikone-R (containing the Accord LTR) expressing nuclear-localized GFP replicate the expression pattern seen by in situ hybridization. (A and B) Expression of GFP in the midgut (MG) and Malpighian tubules (MT) is detected using the Cyp6g1 promoter of Canton-S in third instar larvae. (C and D) Expression of GFP in the midgut (MG) and Malpighian tubules (MT) is detected using the Cyp6g1 promoter of y;cn bw sp. Faint expression of GFP in the fat body was detected only in wandering third instar larvae but not feeding third instar larvae. (E and F) Expression of GFP in the MG, MT, gastric caecae (GC), and the fat body (FB) is detected throughout the third instar larval stage when GFP is driven by the promoter of Hikone-R (containing the Accord LTR).
The Accord element carries tissue-specific enhancers:
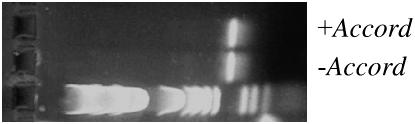
Although there are some single-base differences in the upstream region of Cyp6g1 between y; cn bw sp, Canton-S, and Hikone-R (Daborn et al. 2002), by far the biggest difference is the Accord LTR insertion in Hikone-R. This motivated our characterization of the effect of the Accord LTR insertion on Cyp6g1 expression. It has been reported previously that TEs can carry their own promoters that are capable of changing the transcription start site of the genes adjacent to the TE insertion point (Landry et al. 2002; Dunn et al. 2003). The presence or absence of the Accord LTR upstream of Cyp6g1 did not alter the transcription start site of Cyp6g1 as determined by 5′ RACE (Figure 4). Some studies have shown that TEs can alter the spatial expression of genes they insert near either by disrupting regulatory elements (Lerman and Feder 2004) or by carrying their own enhancers (Conte et al. 2002). To determine if the Accord LTR itself carries an enhancer, we cloned the 491-bp Accord LTR into pH-Stinger, which contains a Hsp70 minimal promoter driving nuclear-localized GFP (Barolo et al. 2000), and transformed w1118 flies with this construct. The expression pattern was the same as that in strains carrying the Accord insertion upstream of Cyp6g1, i.e., expression in the gastric cecum, the entire midgut, Malpighian tubules, and the fat body throughout third instar (Figure 5). This suggests that the Accord LTR carries its own tissue-specific enhancer(s) capable of expressing Cyp6g1 in these tissues. No GFP expression was detected when the Accord LTR was cloned into p-Stinger, which does not contain the Hsp70 minimal promoter (data not shown).
Figure 4.—
5′ RACE performed on Cyp6g1 in flies with and without the Accord LTR shows identical-sized products. This indicates that the Accord LTR does not change the transcription start site of Cyp6g1.
Figure 5.—
Expression pattern of GFP driven by the 491-bp Accord LTR placed in front of the Hsp70 minimal promoter. (A) Expression of GFP is seen in the midgut (MG), Malpighian tubules (MT), and (B) fat bodies (FB) and gastric caecae (GC), indicating that the Accord LTR carries cis-regulatory sequences for expression in these tissues. No GFP expression was detected when the Hsp70 minimal promoter is not present (data not shown).
Expression of Cyp6g1 in the 5′ 6g1-Hikone-R pattern results in insecticide resistance:
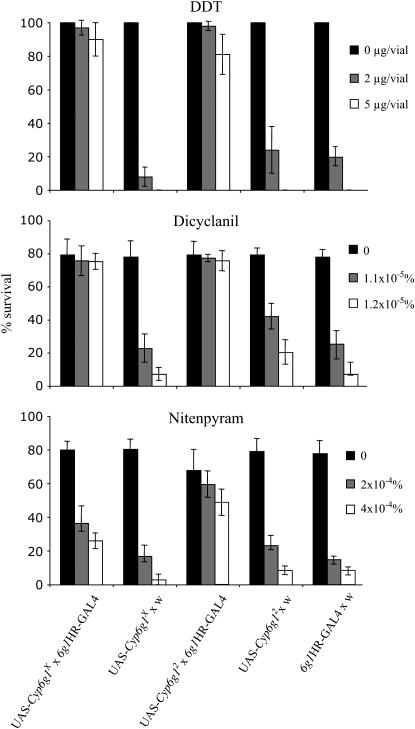
It has previously been shown that the increased expression of Cyp6g1 in D. melanogaster results in resistance to multiple insecticides (Daborn et al. 2001) and that driving the expression of Cyp6g1 ubiquitously using the GAL4/UAS system results in insecticide resistance (Daborn et al. 2002; Le Goff et al. 2003). These studies have made use of GAL4 drivers with a ubiquitous expression pattern, not the exact expression pattern found in resistant strains. To formally link the Accord LTR insertion allele with insecticide resistance, we have used the 6g1HR-GAL4-6c strain to express Cyp6g1 in the same tissues as those in strains carrying the Accord LTR insertion allele. Expressing Cyp6g1 in these tissues does result in resistance for the three insecticides tested (Figure 6), confirming that the Accord LTR insertion upstream of Cyp6g1 is sufficient to confer insecticide resistance. Resistance is seen at both larval and adult life stages. We have presented a detailed characterization of Cyp6g1 expression in larvae and the effect of the Accord LTR. Cyp6g1 expression in adults is increased in Accord-containing strains (Daborn et al. 2002). Both the 6g1HR-GAL4-6c and the pH-Stinger-Accord LTR constructs drive gene expression in similar tissues in adults as in larvae (data not shown), which probably accounts for resistance to DDT at adult life stages.
Figure 6.—
Percentage of survival on discriminating concentrations of the insecticides DDT, dicyclanil, and nitenpyram in two independent UAS-Cyp6g1 lines (UAS-Cyp6g1X where the insert maps to the X chromosome and UAS-Cyp6g12 where the insert maps to the second chromosome). Cyp6g1 is driven in the midgut, Malpighian tubules, and the fat body using the 6g1HR-GAL4-6c strain generated in this study. Increased survival is not seen in controls that do not drive the tissue-specific expression of Cyp6g1 (UAS-Cyp6g1 × w and w × 6g1HR-GAL4-6c). Values are the mean of five replicates ± SEM.
DISCUSSION
In this study, we have documented the native expression pattern of the insecticide resistance gene Cyp6g1 in third instar larvae by in situ hybridization and showed using transgenic constructs that all cis-regulatory elements driving the expression of this gene are located within 1.2 kb of its promoter. We also demonstrated that increased expression of Cyp6g1, which leads to insecticide resistance, is due to the presence of the Accord LTR insertion upstream of Cyp6g1. The Accord LTR does not change the transcription start site of Cyp6g1 but instead carries tissue-specific enhancers that result in Cyp6g1 expression in the Drosophila gastric cecum, midgut, Malpighian tubules, and fat body.
There are an increasing number of examples of TEs contributing to shape the genomes of many organisms (Brookfield 2003; Kazazian 2004), possibly creating new functions for genes (Aminetzach et al. 2005), or contributing novel regulatory sequences (Jordan et al. 2003). The Accord LTR insertion allele of Cyp6g1 has swept to a high frequency in D. melanogaster populations around the world (Catania et al. 2004), indicating that it confers an adaptive phenotype to its host. Our finding that the Accord LTR contains tissue-specific enhancers that function within the host genome is of significance, as there are very few examples where the molecular consequences of TE insertions have been studied in such detail.
In insects, evidence of retrotransposon-derived enhancer sequences that independently regulate the expression of endogenous genes in a tissue-specific manner is limited. One example is the ZAM retrotransposon from D. melanogaster. Cis-acting regulatory sequences in the 5′-untranslated region of ZAM were shown to increase transcription of the white gene (Conte et al. 2002). It is not known if these regulatory sequences are tissue specific, however, as the adult eye was the only tissue where the expression of white was characterized. In another study, an enhancer found in a 17.6 element in D. melanogaster has been characterized that directs the expression in the eye lamina (Mozer and Benzer 1994). This element, however, is not known to regulate the expression of any gene. We provide an example where not only does the TE provide tissue-specific regulatory elements, but these elements actually contribute to a selected phenotype in natural populations, due to its effect on the insecticide resistance-conferring gene, Cyp6g1. The regulatory elements found within the Accord LTR actually independently drive expression in similar tissues to the endogenous Cyp6g1 gene.
TEs have been shown to adapt to the host genome by evolving sequences within them that provide novel regulatory sequences. This is not the case for the Accord LTR in this study. The Accord LTR sequence upstream of Cyp6g1 is identical to the LTR of the canonical Accord elements present in the y; cn bw sp strain (Kaminker et al. 2002). It is therefore likely that the Accord LTR intrinsically carries the regulatory sequences to confer the tissue-specific expression. Other TEs have also been shown to carry tissue-specific enhancers in their LTR or 5′-UTR (Pi et al. 2004; Ruda et al. 2004). Enhancer sequences are presumably involved in the expression of TEs in temporal and tissue-specific patterns as shown by the BDGP embryonic in situ hybridization project (Tomancak et al. 2002). The fact that the Accord LTR carries tissue-specific enhancers for the midgut, Malpighian tubules, and the fat body and has inserted upstream of a gene capable of detoxifying insecticides makes this story remarkable. These tissues play major roles in toxin metabolism and excretion (Hoshizaki 2005; Dow and Davies 2006), although little work has so far been published showing these tissues to directly metabolize insecticides (Suchail et al. 2004).
D. melanogaster is not the only species where a TE has inserted upstream of Cyp6g1. In a striking example of parallel evolution, a Doc element has inserted into the upstream region of the D. simulans Cyp6g1 ortholog and is associated with the overexpression of this gene (Schlenke and Begun 2004). The exact mechanism of D. simulans Cyp6g1 overexpression has thus far not been characterized, although it is likely that Doc-associated sequences affect the expression of the gene. This highlights the importance of increased or tissue-specific expression of Cyp6g1 in providing a selective advantage in natural Drosophila populations and the potential importance of TEs in rapidly providing the suitable molecular changes upon which selection can act.
A recent study has shown that some retrotransposon insertions associated with specific genes are fixed in diverse populations of D. melanogaster (Franchini et al. 2004). There is evidence that the Accord allele has rapidly swept to high frequencies in D. melanogaster populations around the world (Daborn et al. 2002; Catania et al. 2004). It is not clear exactly what has driven this sweep, although selection by insecticides is possible. Our data indicate that the Accord insertion confers resistance to a broad spectrum of widely used chemical insecticides. Drnevich et al. (2004) present evidence indicating that there may be a fitness cost in terms of male reproductive success under competitive conditions in the laboratory. Chemical selection could provide a cogent explanation for the observed sweep of the Accord allele even in the face of selective disadvantage in the field. McCart et al. (2005) have examined a range of life history traits and reported a female reproductive advantage associated with the Accord allele. This advantage may therefore have some capacity to explain the selective sweep as an alternative hypothesis to chemical selection. Clearly further analysis of the fitness of Cyp6g1 genotypes in the field is required, as is a thorough investigation of other possible functions of Cyp6g1. This example indicates that the detailed analysis of other retrotransposon insertions at high frequencies in natural populations may be fruitful in investigating the molecular basis of adaptation.
Acknowledgments
We thank Charles Robin for valuable discussion, Richard Burke for advice on in situ hybridization, and Jon Martin for help with in situ hybridization images. This work was supported by grants from the Australian Research Council through its funding of the Special Research Centre CESAR (Centre for Environmental Stress and Adaptation Research). P.J.D. is supported by an Australian Research Council Linkage–Australian Postdoctoral Fellowship (Commonwealth Scientific and Industrial Research Organisation).
References
- Aminetzach, Y. T., J. M. Macpherson and D. A. Petrov, 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309: 764–767. [DOI] [PubMed] [Google Scholar]
- Argeson, A. C., K. K. Nelson and L. D. Siracusa, 1996. Molecular basis of the pleiotropic phenotype of mice carrying the hypervariable yellow (Ahvy) mutation at the agouti locus. Genetics 142: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo, S., L. A. Carver and J. W. Posakony, 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29: 726, 728, 730, 732. [DOI] [PubMed]
- Bogwitz, M. R., H. Chung, L. Magoc, S. Rigby, W. Wong et al., 2005. Cyp12a4 confers lufenuron resistance in a natural population of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 102: 12807–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Britten, R. J., 1996. Cases of ancient mobile element DNA insertions that now affect gene regulation. Mol. Phylogenet. Evol. 5: 13–17. [DOI] [PubMed] [Google Scholar]
- Britten, R. J., 2004. Coding sequences of functioning human genes derived entirely from mobile element sequences. Proc. Natl. Acad. Sci. USA 101: 16825–16830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield, J. F., 2003. Mobile DNAs: the poacher turned gamekeeper. Curr. Biol. 13: R846–R847. [DOI] [PubMed] [Google Scholar]
- Catania, F., M. O. Kauer, P. J. Daborn, J. L. Yen, R. H. ffrench-Constant et al., 2004. World-wide survey of an Accord insertion and its association with DDT resistance in Drosophila melanogaster. Mol. Ecol. 13: 2491–2504. [DOI] [PubMed] [Google Scholar]
- Conte, C., B. Dastugue and C. Vaury, 2002. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell. Biol. 22: 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn, P., S. Boundy, J. Yen, B. Pittendrigh and R. ffrench-Constant, 2001. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genomics 266: 556–563. [DOI] [PubMed] [Google Scholar]
- Daborn, P. J., J. L. Yen, M. R. Bogwitz, G. Le Goff, E. Feil et al., 2002. A single p450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. [DOI] [PubMed] [Google Scholar]
- Doolittle, W. F., and C. Sapienza, 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601–603. [DOI] [PubMed] [Google Scholar]
- Dow, J. A., and S. A. Davies, 2006. The Malpighian tubule: rapid insights from post-genomic biology. J. Insect. Physiol. 52: 365–378. [DOI] [PubMed] [Google Scholar]
- Drnevich, J. M., M. M. Reedy, E. A. Ruedi, S. Rodriguez-Zas and K. A. Hughes, 2004. Quantitative evolutionary genomics: differential gene expression and male reproductive success in Drosophila melanogaster. Proc. Biol. Sci. 271: 2267–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, C. A., P. Medstrand and D. L. Mager, 2003. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. USA 100: 12841–12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ffrench-Constant, R., P. Daborn and R. Feyereisen, 2006. Resistance and the jumping gene. BioEssays 28: 6–8. [DOI] [PubMed] [Google Scholar]
- Franchini, L. F., E. W. Ganko and J. F. McDonald, 2004. Retrotransposon-gene associations are widespread among D. melanogaster populations. Mol. Biol. Evol. 21: 1323–1331. [DOI] [PubMed] [Google Scholar]
- Gahan, L. J., F. Gould and D. G. Heckel, 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293: 857–860. [DOI] [PubMed] [Google Scholar]
- Gdula, D. A., T. I. Gerasimova and V. G. Corces, 1996. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl. Acad. Sci. USA 93: 9378–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshizaki, D. K., 2005. Fat-cell development, pp. 315–345 in Comprehensive Molecular Insect Science, edited by L. I. Gilbert, K. Iatrou and S. S. Gill. Elsevier, Amsterdam/New York.
- Jordan, I. K., I. B. Rogozin, G. V. Glazko and E. V. Koonin, 2003. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19: 68–72. [DOI] [PubMed] [Google Scholar]
- Kaminker, J. S., C. M. Bergman, B. Kronmiller, J. Carlson, R. Svirskas et al., 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3: RESEARCH0084. [DOI] [PMC free article] [PubMed]
- Kazazian, Jr., H. H., 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Landry, J. R., A. Rouhi, P. Medstrand and D. L. Mager, 2002. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 19: 1934–1942. [DOI] [PubMed] [Google Scholar]
- Le Goff, G., S. Boundy, P. J. Daborn, J. L. Yen, L. Sofer et al., 2003. Microarray analysis of cytochrome P450 mediated insecticide resistance in Drosophila. Insect Biochem. Mol. Biol. 33: 701–708. [DOI] [PubMed] [Google Scholar]
- Lerman, D. N., and M. E. Feder, 2004. Naturally occurring transposable elements disrupt hsp70 promoter function in Drosophila melanogaster. Mol. Biol. Evol. 22(3): 776–783. (erratum: Mol. Biol. Evol. 22(4): 1160). [DOI] [PubMed] [Google Scholar]
- Magoc, L., J. L. Yen, A. Hill-Williams, J. A. McKenzie, P. Batterham et al., 2005. Cross-resistance to dicyclanil in cyromazine-resistant mutants of Drosophila melanogaster and Lucilia cuprina. Pestic. Biochem. Physiol. 81: 129–135. [Google Scholar]
- Marsano, R. M., R. Caizzi, R. Moschetti and N. Junakovic, 2005. Evidence for a functional interaction between the Bari1 transposable element and the cytochrome P450 Cyp12a4 gene in Drosophila melanogaster. Gene 357: 122–128. [DOI] [PubMed] [Google Scholar]
- McCart, C., A. Buckling and R. H. ffrench-Constant, 2005. DDT resistance in flies carries no cost. Curr. Biol. 15: R587–589. [DOI] [PubMed] [Google Scholar]
- McKenzie, J. A., and P. Batterham, 1994. The genetic, molecular and phenotypic consequences of selection for insecticide resistance. Trends Ecol. Evol. 9: 166–169. [DOI] [PubMed] [Google Scholar]
- Mozer, B. A., and S. Benzer, 1994. Ingrowth by photoreceptor axons induces transcription of a retrotransposon in the developing Drosophila brain. Development 120: 1049–1058. [DOI] [PubMed] [Google Scholar]
- Orgel, L. E., and F. H. Crick, 1980. Selfish DNA: the ultimate parasite. Nature 284: 604–607. [DOI] [PubMed] [Google Scholar]
- Parnell, T. J., M. M. Viering, A. Skjesol, C. Helou, E. J. Kuhn et al., 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100: 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi, W., Z. Yang, J. Wang, L. Ruan, X. Yu et al., 2004. The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc. Natl. Acad. Sci. USA 101: 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, F. M., M. R. Bogwitz, T. Perry, A. Monk, P. Batterham et al., 2004. The genetic basis of resistance to diazinon in natural populations of Drosophila melanogaster. Genetica 121: 13–24. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348–353. [DOI] [PubMed] [Google Scholar]
- Ruda, V. M., S. B. Akopov, D. O. Trubetskoy, N. L. Manuylov, A. S. Vetchinova et al., 2004. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res. 104: 11–16. [DOI] [PubMed] [Google Scholar]
- Schlenke, T. A., and D. J. Begun, 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Suchail, S., G. De Sousa, R. Rahmani and L. P. Belzunces, 2004. In vivo distribution and metabolisation of 14C-imidacloprid in different compartments of Apis mellifera L. Pest Manag. Sci. 60: 1056–1062. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81–85. [DOI] [PubMed] [Google Scholar]
- Tomancak, P., A. Beaton, R. Weiszmann, E. Kwan, S. Shu et al., 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T. G., 1993. Transposable elements as initiators of insecticide resistance. J. Econ. Entomol. 86: 645–651. [DOI] [PubMed] [Google Scholar]