Abstract
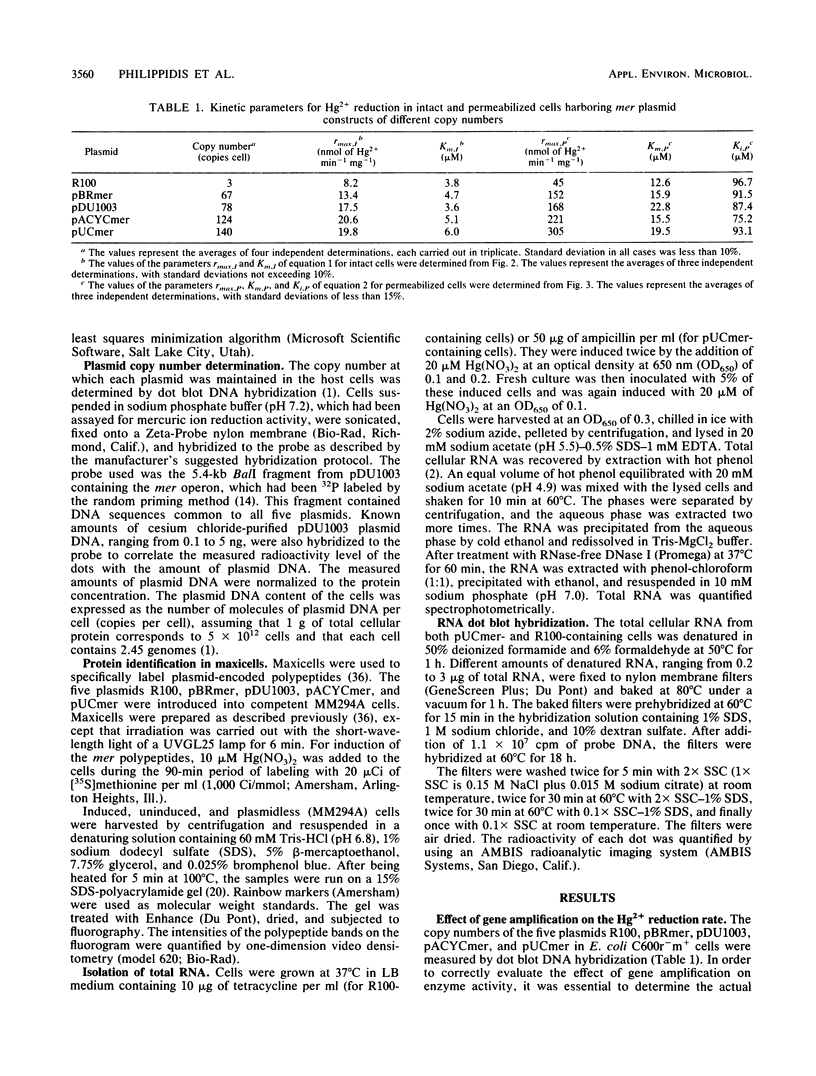
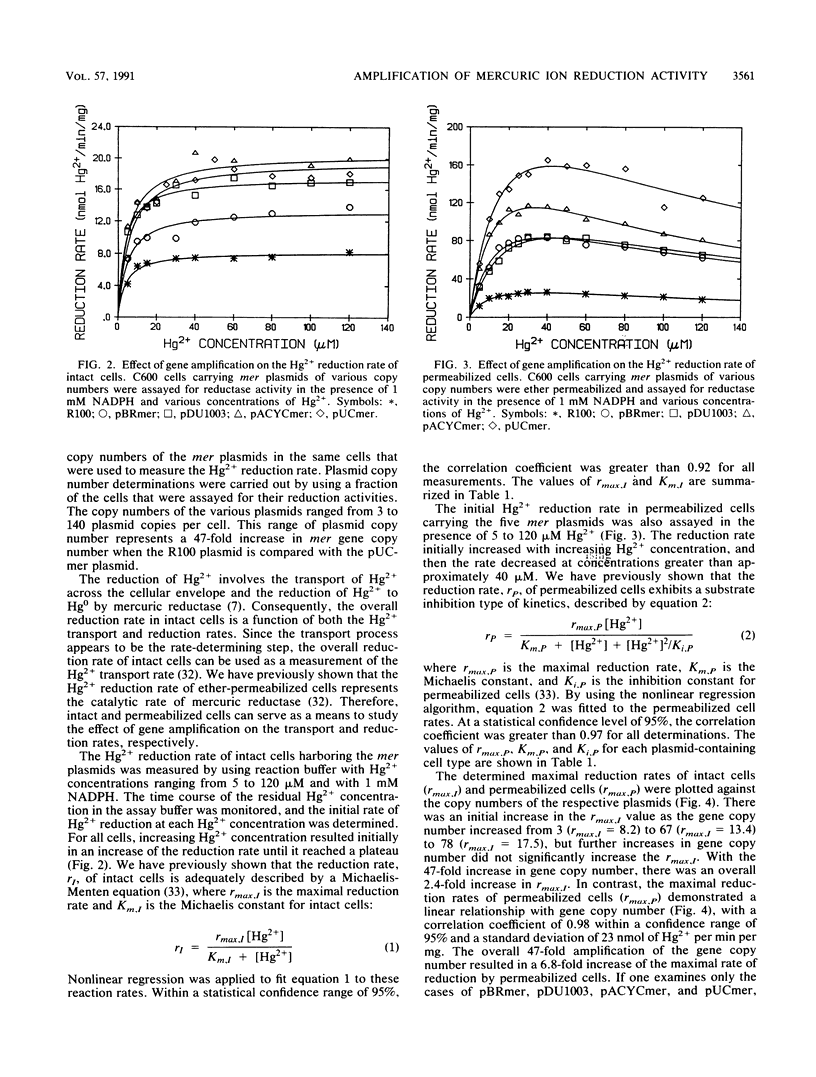
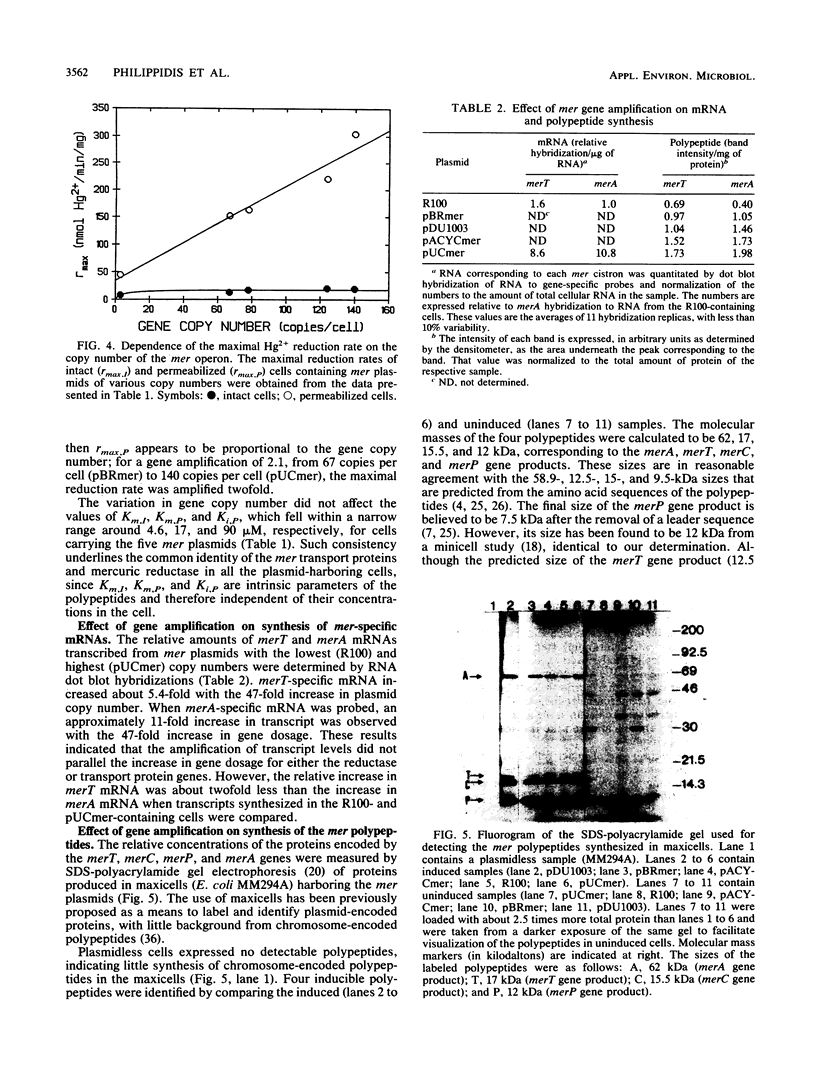
The mercury resistance (mer) operon of plasmid R100 was cloned onto various plasmid vectors to study the effect of mer gene amplification on the rate of Hg2+ reduction by Escherichia coli cells. The plasmids were maintained at copy numbers ranging from 3 to 140 copies per cell. The overall Hg2+ reduction rate of intact cells increased only 2.4-fold for the 47-fold gene amplification. In contrast, the rate of the cytoplasmic reduction reaction, measured in permeabilized cells, increased linearly with increasing gene copy number, resulting in a 6.8-fold overall amplification. RNA hybridizations indicated that mRNA of the cytoplasmic mercuric reductase (merA gene product) increased 11-fold with the 47-fold gene amplification, while mRNA of the transport protein (merT gene product) increased only 5.4-fold. Radiolabeled proteins produced in maxicells were used to correlate the expression levels of the mer polypeptides with the measured reduction rates. The results indicated that, with increasing gene copy number, there was an approximately 5-fold increase in the merA gene product compared with a 2.5-fold increase in the merT gene product. These data demonstrate a parallel increase of Hg2+ reduction activity and transport protein expression in intact cells with plasmids with different copy numbers. In contrast, the expression level of the mercuric reductase gene underwent higher amplification than that of the transport genes at both the RNA and protein levels as plasmid copy number increased.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. W., Hatfield G. W. Effects of promoter strengths and growth conditions on copy number of transcription-fusion vectors. J Biol Chem. 1984 Jun 25;259(12):7399–7403. [PubMed] [Google Scholar]
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Barrineau P., Gilbert P., Jackson W. J., Jones C. S., Summers A. O., Wisdom S. The DNA sequence of the mercury resistance operon of the IncFII plasmid NR1. J Mol Appl Genet. 1984;2(6):601–619. [PubMed] [Google Scholar]
- Bhriain N. N., Foster T. J. Polypeptides specified by the mercuric resistance (mer) operon of plasmid R100. Gene. 1986;42(3):323–330. doi: 10.1016/0378-1119(86)90236-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brown N. L., Misra T. K., Winnie J. N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. 1986 Jan;202(1):143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- Bujard H., Baldari C., Brunner M., Deuschle U., Gentz R., Hughes J., Kammerer W., Stuber D. Integration of efficient promoters of the E. coli system into plasmid vectors. Gene Amplif Anal. 1983;3:65–87. [PubMed] [Google Scholar]
- Chambers S. P., Prior S. E., Barstow D. A., Minton N. P. The pMTL nic- cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988 Aug 15;68(1):139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distefano M. D., Au K. G., Walsh C. T. Mutagenesis of the redox-active disulfide in mercuric ion reductase: catalysis by mutant enzymes restricted to flavin redox chemistry. Biochemistry. 1989 Feb 7;28(3):1168–1183. doi: 10.1021/bi00429a035. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H., Weiss A. A., Silver S. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J Bacteriol. 1979 Oct;140(1):167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Izaki K., Tashiro Y., Funaba T. Mechanism of mercuric chloride resistance in microorganisms. 3. Purification and properties of a mercuric ion reducing enzyme from Escherichia coli bearing R factor. J Biochem. 1974 Mar;75(3):591–599. doi: 10.1093/oxfordjournals.jbchem.a130427. [DOI] [PubMed] [Google Scholar]
- Jackson W. J., Summers A. O. Biochemical characterization of HgCl2-inducible polypeptides encoded by the mer operon of plasmid R100. J Bacteriol. 1982 Aug;151(2):962–970. doi: 10.1128/jb.151.2.962-970.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W. J., Summers A. O. Polypeptides encoded by the mer operon. J Bacteriol. 1982 Feb;149(2):479–487. doi: 10.1128/jb.149.2.479-487.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lund P. A., Brown N. L. Role of the merT and merP gene products of transposon Tn501 in the induction and expression of resistance to mercuric ions. Gene. 1987;52(2-3):207–214. doi: 10.1016/0378-1119(87)90047-3. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Miller S. M., Ballou D. P., Massey V., Williams C. H., Jr, Walsh C. T. Two-electron reduced mercuric reductase binds Hg(II) to the active site dithiol but does not catalyze Hg(II) reduction. J Biol Chem. 1986 Jun 25;261(18):8081–8084. [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Fritzinger D. C., Pridmore R. D., Barnes W. M., Haberstroh L., Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Haberstroh L., Schmidt A., Goddette D., Silver S. Mercuric reductase structural genes from plasmid R100 and transposon Tn501: functional domains of the enzyme. Gene. 1985;34(2-3):253–262. doi: 10.1016/0378-1119(85)90134-9. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Hashimoto H., Mickel S., Rownd R. Round of replication mutant of a drug resistance factor. J Bacteriol. 1974 Jun;118(3):855–866. doi: 10.1128/jb.118.3.855-866.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Kinscherf T. G., Silver S., Miki T., Easton A. M., Rownd R. H. Gene copy number effects in the mer operon of plasmid NR1. J Bacteriol. 1979 Apr;138(1):284–287. doi: 10.1128/jb.138.1.284-287.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni'Bhriain N. N., Silver S., Foster T. J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983 Aug;155(2):690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora G., Silver S., Misra T. K. Down regulation of the mercury resistance operon by the most promoter-distal gene merD. Mol Gen Genet. 1989 Dec;220(1):69–72. doi: 10.1007/BF00260858. [DOI] [PubMed] [Google Scholar]
- Rinderle S. J., Booth J. E., Williams J. W. Mercuric reductase from R-plasmid NR1: characterization and mechanistic study. Biochemistry. 1983 Feb 15;22(4):869–876. doi: 10.1021/bi00273a025. [DOI] [PubMed] [Google Scholar]
- Rose R. E. The nucleotide sequence of pACYC184. Nucleic Acids Res. 1988 Jan 11;16(1):355–355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottel J. L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978 Jun 25;253(12):4341–4349. [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Mercury resistance in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1972 Dec;112(3):1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Teather R. M., Bramhall J., Riede I., Wright J. K., Fürst M., Aichele G., Wilhelm U., Overath P. Lactose carrier protein of Escherichia coli. Structure and expression of plasmids carrying the Y gene of the lac operon. Eur J Biochem. 1980;108(1):223–231. doi: 10.1111/j.1432-1033.1980.tb04715.x. [DOI] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- Vosberg H. P., Hoffmann-Berling H. DNA synthesis in nucleotide-permeable Escherichia coli cells. I. Preparation and properties of ether-treated cells. J Mol Biol. 1971 Jun 28;58(3):739–753. doi: 10.1016/0022-2836(71)90037-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]