Abstract
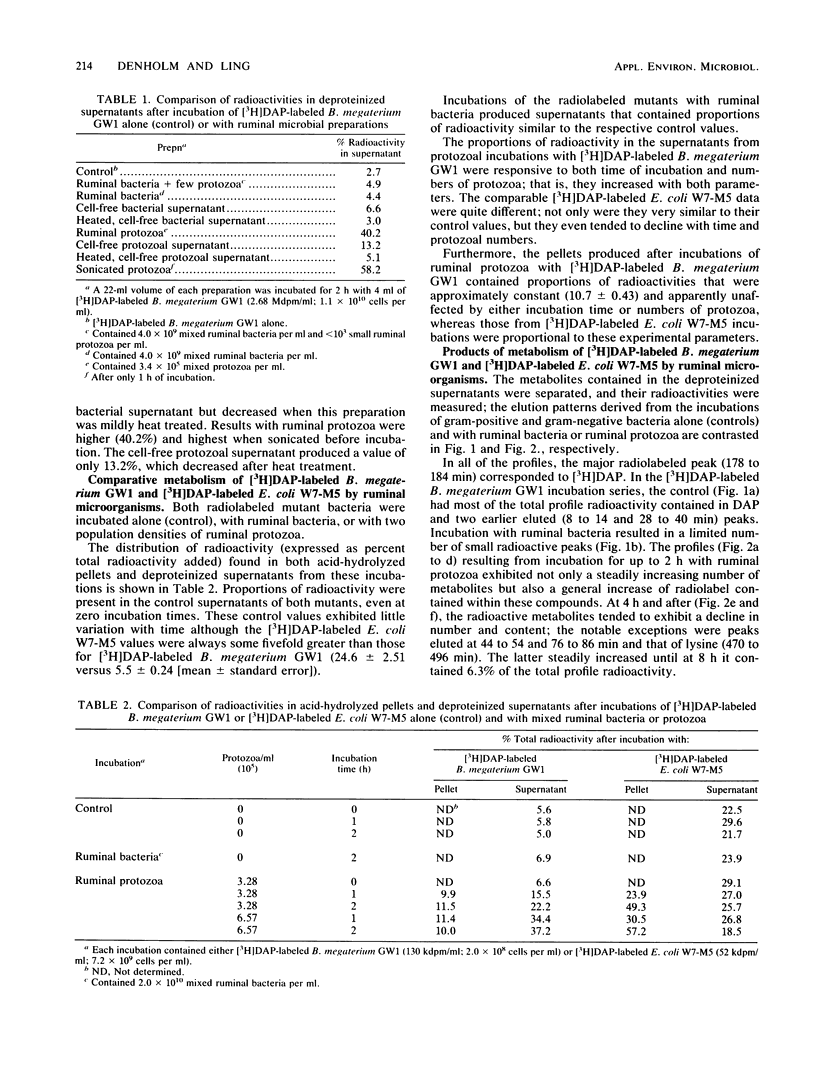
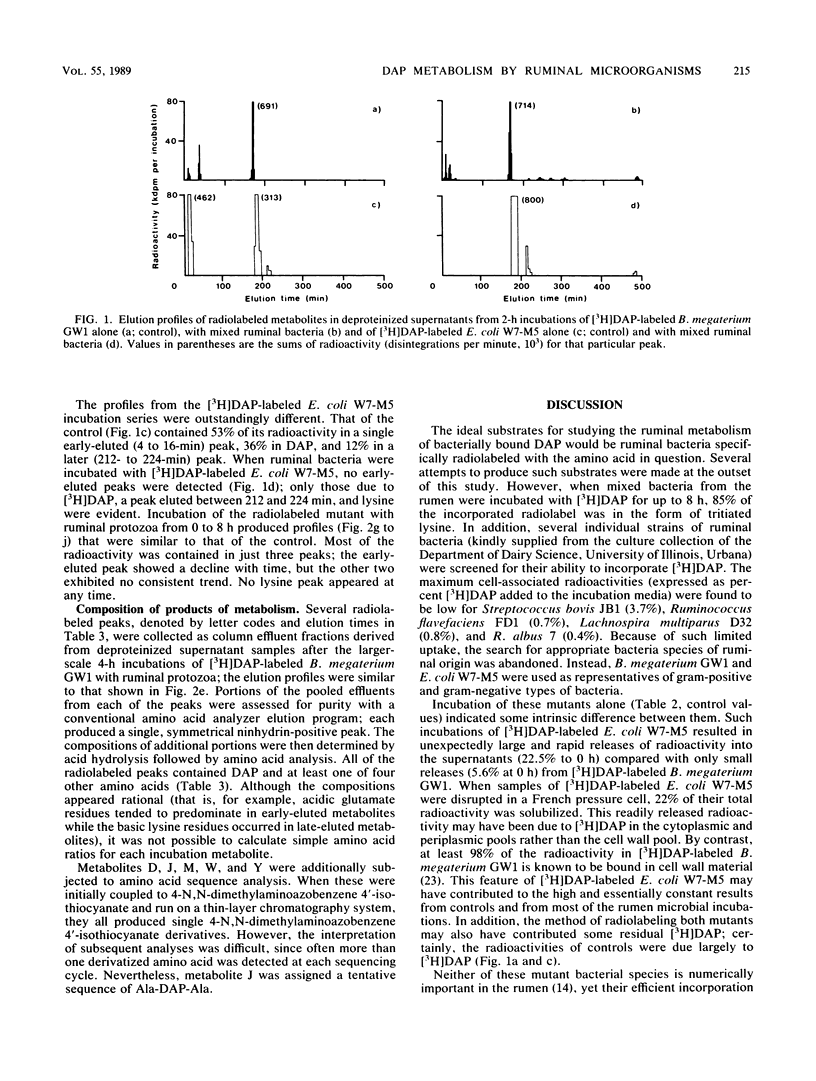
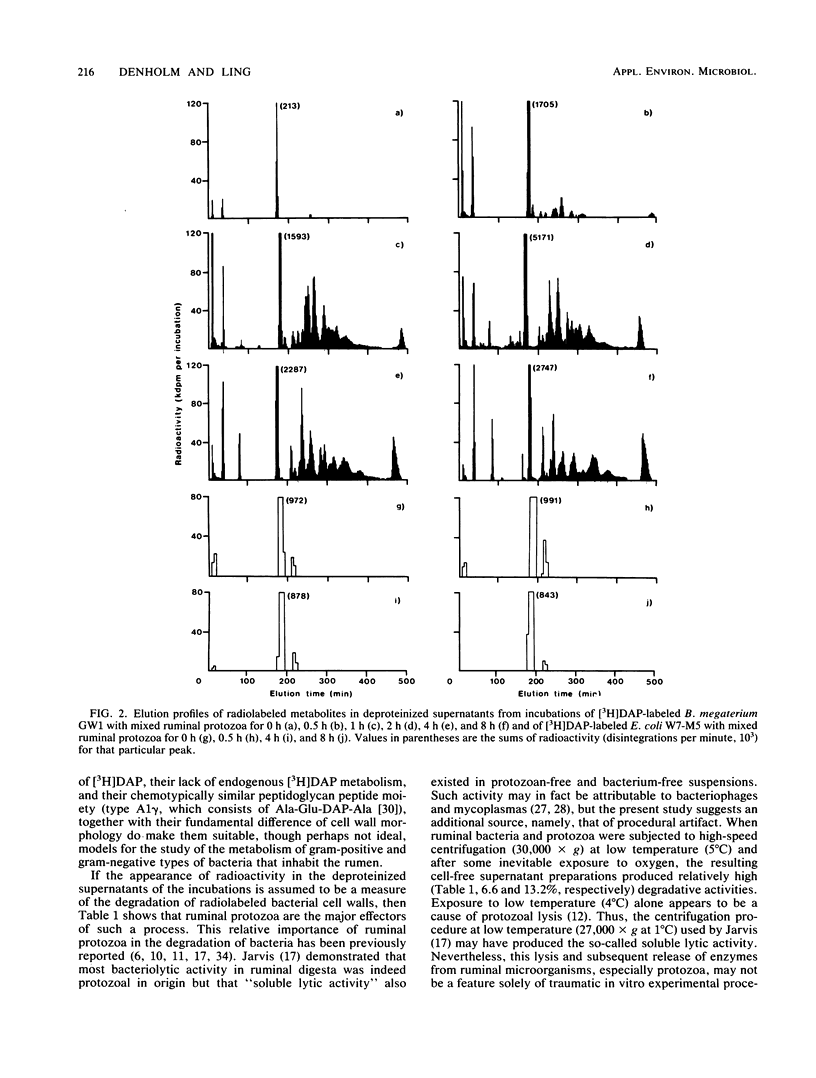
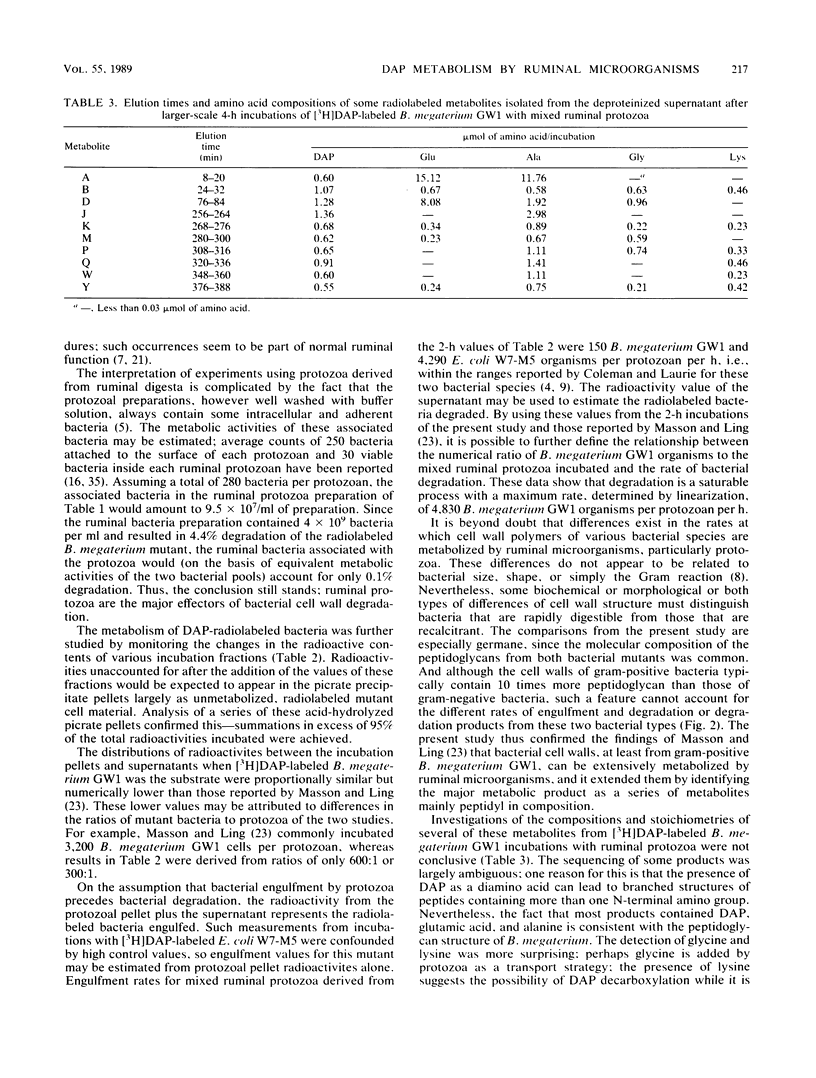
Bacillus megaterium GW1 and Escherichia coli W7-M5 were specifically radiolabeled with 2,2'-diamino[G-3H]pimelic acid [( 3H]DAP) as models of gram-positive and gram-negative bacteria, respectively. These radiolabeled bacterial mutants were incubated alone (control) and with mixed ruminal bacteria or protozoa, and the metabolic processes, rates, and patterns of radiolabeled products released from them were studied. Control incubations revealed an inherent difference between the two substrates; gram-positive supernatants consistently contained 5% radioactivity, whereas even at 0 h, those from the gram-negative mutant released 22%. Incubations with ruminal microorganisms showed that the two mutants were metabolized differently and that protozoa were the major effectors of their metabolism. Protozoa exhibited differential rates of engulfment (150 B. megaterium GW1 and 4,290 E. coli W7-M5 organisms per protozoan per h), and they extensively degraded [3H]DAP-labeled B. megaterium GW1 at rates up to nine times greater than those of ruminal bacteria. By contrast, [3H]DAP-labeled E. coli W7-M5 degradation by either ruminal bacteria or ruminal protozoa was more limited. These fundamental differences in the metabolism of the two mutants, especially by ruminal protozoa, were reflected in the patterns and rates of radiolabeled metabolites produced; many were rapidly released from [3H]DAP-labeled B. megaterium GW1, whereas few were slowly released from [3H]DAP-labeled E. coli W7-M5. Most radiolabeled products derived from [3H]DAP-labeled B. megaterium GW1 were peptides of bacterial peptidoglycan origin. The ruminal metabolism of DAP-containing gram-positive and gram-negative bacteria, even with the same peptidoglycan chemotype, is thus likely to be profoundly different.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coleman G. S., Laurie J. I. The utilization of Bacillus megaterium and the release of a lytic enzyme by three Epidinium spp. isolated from the rumen. J Gen Microbiol. 1974 Dec;85(2):257–264. doi: 10.1099/00221287-85-2-257. [DOI] [PubMed] [Google Scholar]
- Coleman G. S. Rumen ciliate protozoa. Adv Parasitol. 1980;18:121–173. doi: 10.1016/s0065-308x(08)60399-1. [DOI] [PubMed] [Google Scholar]
- Forsberg C. W., Lovelock L. K., Krumholz L., Buchanan-Smith J. G. Protease activities of rumen protozoa. Appl Environ Microbiol. 1984 Jan;47(1):101–110. doi: 10.1128/aem.47.1.101-110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton K., Bailey F. J., Annison E. F. Measurement of the bacterial nitrogen entering the duodenum of the ruminant using diaminopimelic acid as a marker. Br J Nutr. 1971 Jan;25(1):165–173. doi: 10.1079/bjn19710074. [DOI] [PubMed] [Google Scholar]
- Imai S., Ogimoto K. Scanning electron and fluorescent microscopic studies on the attachment of spherical bacteria to ciliate Protozoa in the ovine rumen. Nihon Juigaku Zasshi. 1978 Feb;40(1):9–19. doi: 10.1292/jvms1939.40.9. [DOI] [PubMed] [Google Scholar]
- Jarvis B. D. Lysis of viable rumen bacteria in bovine rumen fluid. Appl Microbiol. 1968 May;16(5):714–723. doi: 10.1128/am.16.5.714-723.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng R. A. Dynamics of protozoa in the rumen of sheep. Br J Nutr. 1982 Sep;48(2):399–415. doi: 10.1079/bjn19820123. [DOI] [PubMed] [Google Scholar]
- Ling J. R., Buttery P. J. The simultaneous use of ribonucleic acid, 35S, 2,6-diaminopimelic acid and 2-aminoethylphosphonic acid as markers of microbial nitrogen entering the duodenum of sheep. Br J Nutr. 1978 Jan;39(1):165–179. doi: 10.1079/bjn19780023. [DOI] [PubMed] [Google Scholar]
- Orpin C. G., Munn E. A. The occurrence of bacteriophages in the rumen and their influence on rumen bacterial populations. Experientia. 1974 Sep 15;30(9):1018–1020. doi: 10.1007/BF01938983. [DOI] [PubMed] [Google Scholar]
- Schleifer K. H., Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972 Dec;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F., Rittenberg S. C. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: solubilization of Escherichia coli peptidoglycan. J Bacteriol. 1978 Sep;135(3):998–1007. doi: 10.1128/jb.135.3.998-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. J., McPherson C. A. Factors affecting the rate of breakdown of bacterial protein in rumen fluid. Br J Nutr. 1987 Sep;58(2):313–323. doi: 10.1079/bjn19870098. [DOI] [PubMed] [Google Scholar]
- White R. W. Viable bacteria inside the rumen ciliate Entodinium caudatum. J Gen Microbiol. 1969 Jun;56(3):403–408. doi: 10.1099/00221287-56-3-403. [DOI] [PubMed] [Google Scholar]
- Whitelaw F. G., Eadie J. M., Bruce L. A., Shand W. J. Microbial protein synthesis in cattle given roughage-concentrate and all-concentrate diets: the use of 2,6-diaminopimelic acid, 2-aminoethylphosphonic acid and 35S as markers. Br J Nutr. 1984 Sep;52(2):249–260. doi: 10.1079/bjn19840093. [DOI] [PubMed] [Google Scholar]


