Abstract
We have identified six new aminoacylation determinants of Escherichia coli tRNAGln in a genetic and biochemical analysis of suppressor tRNA. The new determinants occupy the interior of the acceptor stem, the inside corner of the L shape, and the anticodon loop of the molecule. They supplement the primary determinants located in the anticodon and acceptor end of tRNAGln described previously. Remarkably, the three-dimensional structure of the complex between tRNAGln and glutaminyl-tRNA synthetase shows that the enzyme interacts with the phosphate–sugar backbone but not the base of every new determinant. Moreover, a small protein motif interacts with five of these determinants, and it binds proximal to the sixth. The motif also interacts with the middle base of the anticodon and with the backbones of six other nucleotides. Our results emphasize that synthetase recognition of tRNA is more elaborate than amino acid side chains of the enzyme interacting with nucleotide bases of the tRNA. Recognition also includes synthetase interaction with tRNA backbone functionalities whose distinctive locations in three-dimensional space are exquisitely determined by the tRNA sequence.
The distinctive sites in tRNA that govern interaction with the correct aminoacyl-tRNA synthetase (aaRS) during amino acid attachment are known in remarkable detail from biochemical and crystallographic studies of tRNA–aaRS complexes (1). Typically, 20% of the accessible surface of tRNA interacts with the aaRS, and some interactions are accompanied by dramatic tRNA conformational changes. It is striking that tRNA backbone-mediated interactions with aaRS are prevalent (2–5), but little is known about the contribution of these interactions to aaRS recognition or the rules by which the nucleotide sequence of the tRNA constrains backbone geometry. Here we describe several tRNA backbone-mediated interactions between Escherichia coli tRNAGln and glutaminyl-tRNA synthetase (GlnRS) that are critical to aminoacylation.
The three-dimensional structure at 2.5-Å resolution of the complex between tRNAGln, GlnRS, and ATP (2, 6), together with genetic and biochemical results (7–9), establish that tRNA recognition relies on enzyme-binding nucleotides in the acceptor end, the interior base pair of the D stem, and anticodon loop of the tRNA. Binding causes extensive conformational changes in the acceptor and anticodon regions of tRNAGln, and the tRNA contains sequences to favor low-energy conformational changes. Importantly, 33% of all nucleotides in tRNAGln are involved in backbone-mediated interactions with the protein.
The results of the above studies allow us to ask which tRNAGln backbone-mediated interactions contribute to aminoacylation. The amber-suppressor tRNA system is appropriate to study tRNAGln because the suppressor anticodon retains U in the middle position (U35), which is a key tRNAGln determinant. Our genetic and biochemical analysis of suppressor tRNA reported below identifies six new determinants that occupy the interior of the acceptor stem (G4⋅C68 and G5⋅C68), the inside corner of the L shape (C11⋅G24, A13-A22, and G15-C48), and the anticodon loop (U32-Ψ38) of the tRNA. The U32-Ψ38 determinant was predicted from the three-dimensional structure of the complex (6), but the other determinants reported here have not been described previously aside from a preliminary description of this project (10). Remarkably, the three-dimensional structure of the tRNAGln–GlnRS complex shows that the enzyme interacts with the phosphate–sugar backbone of every new determinant.
RESULTS AND DISCUSSION
Strategy.
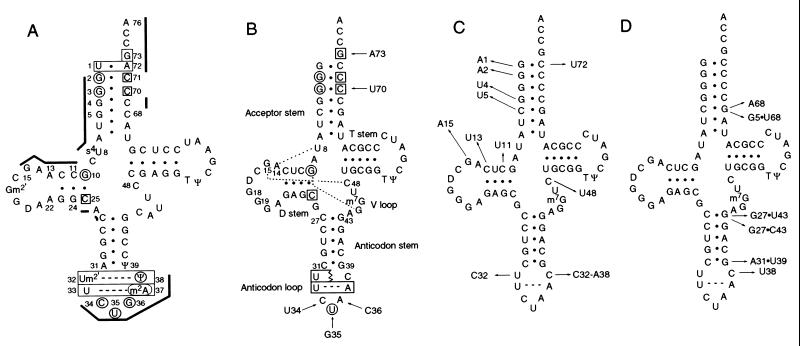
We altered a gene for amber-suppressor tRNAAla so the transcribed tRNA would preserve its native tertiary folding but be inactive with the alanine enzyme and weakly active with the GlnRS enzyme. Fig. 1A shows the cloverleaf arrangement of tRNAGln with annotations of nucleotides that contribute to aminoacylation and/or interact with GlnRS (6). We altered tRNAAla by substituting C70, G73, and the CUA amber-suppressor anticodon in its sequence (Fig. 1B). The C70 and G73 substitutions remove tRNAAla determinants (G3⋅U70, in particular) and generate an acceptor stem that contains all of the known tRNAGln determinants except the U1⋅A72 base pair, which we excluded to impair tRNA activity.
Figure 1.
Cloverleaf arrangement of tRNAs corresponding to tRNAGln (A), the starting tRNA with arrows pointing from the bases of wild-type tRNAAla (UGC anticodon) that were substituted (B), mutant tRNAs with arrows pointing to mutations that improve aminoacylation (C), or mutant tRNAs with arrows pointing to mutations that improve ribosomal performance but do not alter aminoacylation (D). In A, nucleotides that determine aminoacylation of tRNAGln (6, 7, 9) are noted by circles for bases that are directly recognized by GlnRS and boxes for bases that facilitate the tRNA in assuming the conformation bound by the protein; thick lines show phosphate-sugar backbone-mediated interactions with GlnRS. In B, the dashed lines indicate three tertiary pairings discussed in the text. Modified nucleotides were not analyzed.
Cells expressing the inactive suppressor tRNA and containing a lac amber mutation form white colonies on an indicator plate containing 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal). A genetic screen for blue colonies coupled with analyses described below exclude compensating mutations in GlnRS and identify mutations that make the starting tRNA a better GlnRS substrate. Because a transition-type mutagen was used, the compensating mutations are single-base-pair changes in the DNA (G⋅C to A⋅T) that result in wobble base pairs in helical or paired regions of tRNA structure. Compensating mutations that generate wobble base pairs are expected to identify nucleotides in tRNA whose conformations rather than base identities contribute to tRNA aminoacylation. Although wobble base pairs influence RNA conformation in ways that are not completely understood, these pairs are able to influence the location of base and backbone functionalities, ion binding, and helix stability (11–13).
The functional characterization of tRNA mutants relied on freshly transformed clones and cultures in balanced growth (14, 15) and included measuring their suppressor activity by enzyme assay, determining their aminoacylation specificity by sequencing a reporter protein, assessing their aminoacylation efficiency from cellular levels of glutaminyl-tRNA, and measuring their cellular levels of GlnRS to test the possibility that enzyme overproduction is responsible for increases in tRNA aminoacylation. Finally, a mutant tRNA with a defect in protein synthesis was tested for ability to form a stable ternary complex with elongation factor Tu (EF-Tu) and GTP.
Hydroxylamine Mutagenesis.
Many blue colonies were observed on X-gal plates after hydroxylamine mutagenesis. DNA sequencing revealed eight different single-base transitions in the respective tRNAs, each characteristic of the mutagen (Fig. 1C). Three compensating mutations altered nucleotides in the end of the amino acid acceptor stem, one altered the interior of the acceptor stem, two changed the D stem, and the final two changed both nucleotides of the 15–48 tertiary interaction. A representative of each mutant was recloned by restriction-enzyme digestion into fresh plasmid before the functional analysis.
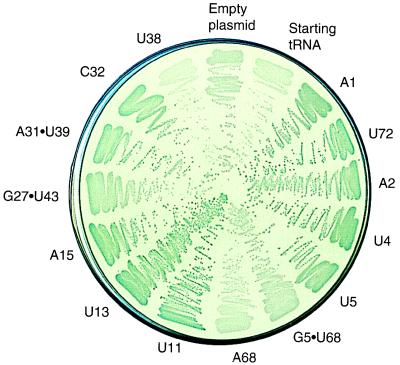
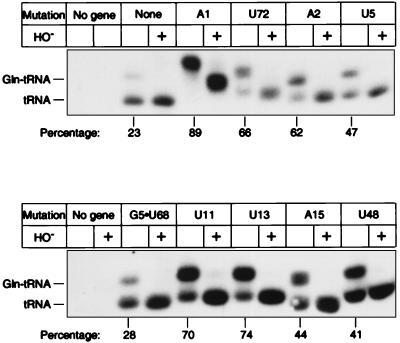
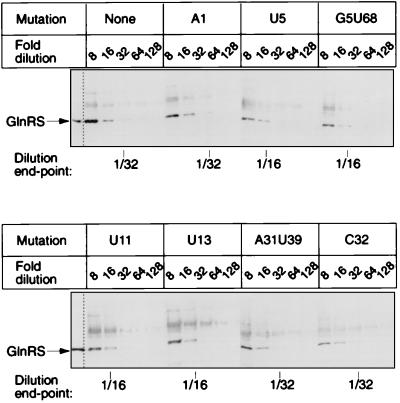
Fig. 2 shows the range of colors on an X-gal plate produced by tRNAs carrying different mutations. Quantitative suppression efficiency measurements in liquid cultures ranged from 0.6 to 8.0% compared with 0.2% for starting tRNA (Table 1). The X-gal plate color and numerical suppression efficiency data are concordant even though the cells in the plate assay are in many growth states, whereas those in liquid cultures are in balanced growth. Suppression efficiency reflects both aminoacylation efficiency and the ribosomal performance (e.g., EF-Tu binding, release factor competition, codon binding) of the tRNA. To examine only aminoacylation, we determined the steady-state levels of glutaminyl-tRNA and uncharged tRNA of cells by using a quantitative Northern tRNA blot analysis. The hybridization probe includes the suppressor anticodon without overlapping compensating mutations and thus binds the suppressor tRNA in preference to chromosomal tRNAs. Sequence determination of the reporter protein showed that each mutant tRNA inserts only Gln (Table 1). The mutant tRNAs are substantially aminoacylated, with steady-state levels ranging from 40 to 90% compared with 23% for the starting suppressor tRNA (Fig. 3 and Table 1). For comparison, we tested amber- suppressor tRNAGln (CUG anticodon isoacceptor) and observed a 22% suppression efficiency and 74% glutaminyl-tRNA. The steady-state level of glutaminyl-tRNA of a mutant tRNA can increase relative to that of the starting tRNA if either the mutant tRNA is a better substrate for GlnRS or if cells expressing the mutant tRNA are overproducing the GlnRS. A Western blot analysis (16) of GlnRS demonstrated that the enzyme level was unchanged in every mutant (Fig. 4 and Table 1), indicating that the mutant tRNAs are better substrates for GlnRS. We next consider how the mutations might work.
Figure 2.
Activity of tRNA mutants on an X-gal indicator plate at 37°C.
Table 1.
Functional properties of tRNA mutants
| Change to starting tRNA | Suppression efficiency, % | Specificity Gln, % | Glutaminyl-tRNA, % | Relative GlnRS |
|---|---|---|---|---|
| None | 0.2 | 96 | 23 | 1.0 |
| Acceptor stem mutations | ||||
| A1 | 0.6 | 95 | 89 | 1.0 |
| U72 | 4.5 | 97 | 66 | 1.0 |
| A2 | 3.2 | 96 | 62 | 1.0 |
| U4 | 0.8 | 92 | 45 | 0.5 |
| U5 | 0.6 | 96 | 47 | 0.5 |
| G5⋅U68* | 0.2 | 96 | 28 | 0.5 |
| A68* | <0.2 | ND | 17 | ND |
| D stem and loop mutations | ||||
| U11 | 8.0 | Unstable | 70 | 0.5 |
| U13 | 2.9 | 95 | 74 | 0.5 |
| A15 | 2.1 | 94 | 44 | 2.0 |
| U48 | 1.4 | 96 | 41 | 0.5 |
| Anticodon stem and loop mutations | ||||
| G27⋅U43* | 1.0 | 94 | 13 | 0.5 |
| G27⋅C43* | 1.1 | 95 | 16 | 1.0 |
| A31⋅U39* | 6.6 | 93 | 23 | 1.0 |
| C32* | 3.9 | 93 | 54 | 1.0 |
| C32-A38* | 24 | 95 | 49 | 1.0 |
| U38* | <0.2 | ND | 17 | 1.0 |
The gene for the starting tRNA was constructed by directed mutagenesis of an E. coli tRNAAla (UGC anticodon) gene in plasmid pGFIB so that the transcribed molecule would contain C34, U35, A36, C70, and G73. Hydroxylamine and directed mutagenesis were used to isolate active mutants forming blue colonies on X-gal indicator plates (Luria–Bertani agar containing ampicillin and X-gal). Suppression efficiency values are the percentage of the wild-type lacI-Z40 fusion, which averaged 138 units. The value for XAC/A16 cells without suppressor tRNA gene was <0.01%. The amino acid specificity is for residue 3 of dihydrofolate reductase protein, except that residue 10 was analyzed for mutant U4; residue 10 also was analyzed for the starting tRNA, which gave results comparable to residue 3 reported above. The U11 mutant was unstable and its protein could not be analyzed. For Northern blot analysis of glutaminyl-tRNA, mutants were analyzed an average of 3.5 times (average SD ±3.1%). The glutaminyl-tRNA values are calculated as [100 × (aminoacyl-tRNA)/(aminoacyl-tRNA + uncharged tRNA)]. For Western blot analysis of GlnRS, mutants were analyzed an average of 3.1 times (reproducibility of ± 2-fold). The values reported are dilution end-points relative to that of the starting tRNA.
*Mutant constructed by directed mutagenesis.
Figure 3.
Northern blot analysis showing aminoacyl-tRNA and uncharged tRNA of representative mutants. tRNA genes with the indicated mutations were expressed from plasmid pGFIB in XAC/A16 cells. Samples containing ≈0.05 OD260 units of total tRNA isolated at pH 5.2 were fractionated by 6.5% PAGE in sodium acetate and urea. Percentage glutaminyl-tRNA is the average of several determinations (Table 1). We verified the glutaminyl-tRNA species by observing its signal increase from 22 to 51% when GlnRS was overproduced from plasmid pAC1.
Figure 4.
Western blot analysis of GlnRS from cells expressing representative mutant tRNAs. Serial dilutions of crude extracts were fractionated by 6% SDS/PAGE. “None” means XAC/A16 cells carrying plasmid pGFIB without an amber-suppressor tRNA gene. The position of purified GlnRS marker is noted. Cells overproducing GlnRS from plasmid pAC1 showed a 4- to 8-fold increase in GlnRS signal.
Mutations at the end of the amino acid acceptor stem can be understood by using the three-dimensional structure of the complex between tRNAGln and GlnRS (2). The enzyme interacts with the backbone of A72 and disrupts the U1⋅A72 base pair by inserting a leucine side-chain between these bases. The A1 and U72 mutations change the G1⋅C72 base pair of the starting tRNA to wobble pairs that facilitate low-energy disruption. The same mutations (and A2 discussed below) (17) have been described in suppressor tRNATyr, where they probably promote aminoacylation in a similar way. Interestingly, the A1 mutant tRNA reported here has a lower suppression efficiency but a higher level of glutaminyl-tRNA than that of other mutants. This result suggests that A1 glutaminyl-tRNA is defective in protein synthesis and accumulates in cells rather than reaching a steady-state level between formation and consumption of glutaminyl-tRNA observed for other mutants.
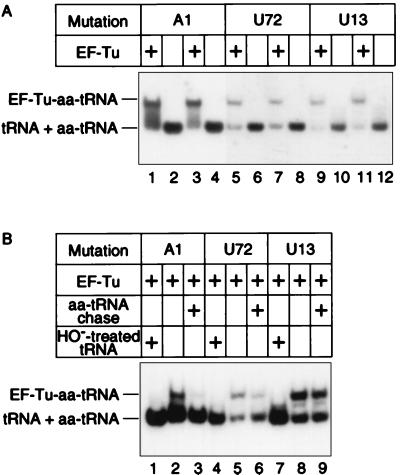
To investigate the A1 defect, we used a gel-retardation assay of the ternary complex between glutaminyl-tRNA and EF-Tu-GTP. Fig. 5A shows that the A1 tRNA and two reference tRNAs form ternary complexes. However, only A1 displays a heterogeneous smear, suggesting that this complex is unstable and dissociates during gel electrophoresis. Fig. 5B shows that A1 glutaminyl-tRNA is not tightly bound because it chases into the unbound species when challenged by competitor aminoacyl-tRNA. The ternary complex of U72 glutaminyl-tRNA is partially chased by competitor aminoacyl-tRNA whereas that of U13 is stable to challenge. The instability of the A1 complex is consistent with other results, including the structure–function relationship of the 1⋅72 pair of elongator and initiator tRNAs with EF-Tu and the crystal structure of the ternary complex (18, 19). In conclusion, EF-Tu binds A1 glutaminyl-tRNA poorly, which curtails the tRNA’s participation in protein synthesis, lowers its suppression efficiency, and causes a partial accumulation of charged tRNA.
Figure 5.
Gel-retardation assay of glutaminyl-tRNA, EF-Tu, and GTP ternary complex. In A, binding mixtures containing 10 μM of an acid preparation of total tRNA, 10 μM EF-Tu-GTP in 46 mM Tris⋅HCl, pH 7.5, 46 mM KCl, 42 mM NH4Cl, 6 mM MgCl2, and 3 mM 2-mercaptoethanol were incubated 5 min at 4°C and fractionated by 6% PAGE. In B, after incubating mixtures as in A, one portion was loaded on the gel being run at 20 V and the other portion was challenged with competitor aminoacyl-tRNA (19 μM final concentration) and incubated another 10 min before loading. The competitor aminoacyl-tRNA was isolated from XAC/A16 cells carrying pGFIB without an amber- suppressor tRNA gene. EF-Tu-GDP was converted to EF-Tu-GTP just before use. Homogeneous preparations of EF-Tu were from either T. aquaticus (A, lanes 1, 2, 5, 6, 9, 10, and B) or T. thermophilus (A, lanes 3, 4, 7, 8, 11, 12). Samples were fractionated at 4°C by 6% PAGE in 25 mM Tris⋅Oac, pH 6.8, 5 mM Mg-Oac, and 5 mM NH4-Oac, transferred by electroblot and hybridized. The gray input levels were adjusted in photoshop 4.0 from 1.00 to 2.00 in A, lanes 1–4, to better compare them with A, lanes 5–12.
The mechanism associated with the enhanced acceptor activity of the A2 tRNA is less certain because GlnRS normally interacts with both the base and the backbone of G2 in tRNAGln (Fig. 1A) while preserving the G2⋅C71 base pairing. The A2⋅C71 wobble pair might facilitate disruption of G1⋅C72 and/or it might adjust local geometry for more favorable enzyme binding. None of the remaining five mutations of the starting tRNA have been identified previously as aminoacylation determinants of tRNAGln.
The U5 mutation creates a U5⋅G68 wobble pair in the interior of the tRNA acceptor stem. In the structure of the wild-type complex, the enzyme interacts with the backbone but not the base of nucleotide 5 of the G5⋅C68 base pair. To consider the U5 mutation further, we constructed tRNAs with G5⋅U68 and C5⋅A68 wobble pairs (Fig. 1D). However, both tRNAs are inactive (Table 1), underscoring the importance of the interior of the acceptor stem for aminoacylation. U5 might enhance aminoacylation by shifting the orientation of base and/or backbone functionalities, or by weakening this part of the helix so the enzyme can more easily modulate it, or by some combination of these possibilities. Modulation of tRNA structure is consistent with the observed displacement of the 5′ acceptor strand of tRNAGln (nucleotides 1 through 7) in the enzyme complex (2).
The remaining mutations map to either the D stem (U11 and U13), the D loop (A15), or the V loop (U48). The enzyme interacts with only the backbone of these residues in tRNAGln (the interaction with C48 is through G15-C48). This group of mutations might improve aminoacylation by weakening tRNA structure as just described for U5, especially because tRNAGln and the starting tRNA contain different numbers of base pairs in the D stem (Fig. 1 A and B). The base pairing on the D stem as well as the nucleotide residues that flank it tend to be semiconserved or conserved in different tRNAs, suggesting that although the D stem and flanking nucleotides alone are not sufficient to discriminate between tRNAs accepting different amino acids, the nucleotides nevertheless are important for aminoacylation.
Directed Mutagenesis.
Our library of hydroxylamine-induced mutations does not include alterations of the anticodon loop of the starting tRNA, even though this part of tRNAGln binds the enzyme. Either mutations in the anticodon loop are unable to improve aminoacylation, or mutations that improve aminoacylation are not generated by hydroxylamine mutagenesis because of the type or number of nucleotide changes required.
To address this question, we used directed mutagenesis. First, C38 was changed to U38 in the transcribed tRNA to re-create the U32–U38 pair of tRNAGln. However, U38 had no effect on tRNA function (Table 1). We also used a mutagenic oligodeoxynucleotide with multiple base mixes at both positions 32 and 38; this gave mutants C32 and C32-A38 with increases in the steady-state level of glutaminyl-tRNA (Table 1). These mutations might weaken the local structure and/or approximate structural features associated with Ψ38 in tRNAGln, including specific hydration of Ψ38 and a small shift in the location of P37 (20). Although the two mutants have similar glutaminyl-tRNA levels, C32-A38 shows a significantly higher suppression efficiency. This finding can be explained by the extended anticodon hypothesis (21), which correlates the A36A37A38 sequence (present only in C32-A38) in suppressor tRNAs with enhanced ribosomal function.
We also mutagenized all of the base pairs on the anticodon stem and T stem to identify mutations able to confer activity on the starting tRNA. (The base pair adjacent to the T loop was not probed.) For this analysis, individual base pairs were separately mutagenized with an oligodeoxynucleotide containing multiple base mixes. This identified three active mutants in the anticodon stem: G27⋅U43 and G27⋅C43 at the first base pair and A31⋅U39 at the last base pair. However, although these mutations increase tRNA suppression efficiency, they do not improve aminoacylation (Table 1). The role of the first and last base pairs of the anticodon stem in influencing ribosomal performance rather than aminoacylation efficiency of suppressor tRNA is well documented (22, 23). Finally, we separately mutagenized the 4⋅69 and 7⋅66 base pairs. This gave one mutant, U4, which creates a U4–C69 pair and improves aminoacylation of the starting tRNA. U4 probably facilitates a backbone-mediated interaction of residue 69 with GlnRS that is important for aminoacylation (see Table 2).
Table 2.
Intermolecular contacts
| Mutation | Nucleotide | Atom | Amino acid | Atom | Distance, Å |
|---|---|---|---|---|---|
| Yes | G5 | O3′ | Lys-317 | NZ | 3.72 |
| U6 | O2P | Lys-317 | CD | 3.09 | |
| U8 | O1P | Gln-318 | OE1 | 2.80 | |
| Yes | C11 | O2′ | Thr-321 | O | 2.86 |
| C12 | O2P | Thr-321 | N | 2.89 | |
| Yes | A13 | O1P | Gln-318 | NE2 | 3.29 |
| A14 | O1P | Thr-316 | CG2 | 3.41* | |
| C25 | O2′ | Ser-326 | N | 3.60 | |
| U35 | O4 | Arg-341 | NH2 | 2.72 | |
| Yes | Ψ38 | O2′ | Asn-336 | ND2 | 3.32 |
| Yes | C69 | O2′ | Asp-319 | OD1 | 3.63 |
Direct contacts between the backbone of tRNAGln and GlnRS observed at 2.5 Å resolution are listed (6). Other interactions are: U6 C4′, Lys-317 O, 3.43 Å; G10 N2, Glu-323 CD, 3.53 Å; G10 N2, Glu-323 OE1, 2.76 Å; C11 O2′, Ile-322 CA, 3.66 Å; C11 O3′, Thr-321 O, 3.09 Å; C11 O4′, Glu323 CD, 3.42 Å; C11 O4′, Glu323 OE2, 3.37Å; C12 O2P, Asn-320 CA, 3.35 Å; C25 O2′, Ala-325 C, 3.63 Å, U38 C4′, Asn-336 OD1, 3.53 Å; U38 O3′, Asn-336 ND2, 3.53 Å. Also, G15 interacts with Gln13 on the N-terminal helix (O1P-NE2, 3.83 Å) and Gln-13 interacts with Phe-10 on the same face of the helix (four distances, 3.30–3.88 Å). Phe-10 packs against the side chains of Val-315 and Thr-316 (six distances, 3.67–3.90 Å).
Assumes a possible rotation of the side chain of Thr-316. The nonrotated distance is 3.84 Å.
Determination of the steady-state level of glutaminyl-tRNA has allowed us to deconvolute tRNA suppression efficiency into its component parts, tRNA aminoacylation efficiency and ribosomal performance. This disclosed that aminoacylation efficiency correlates with suppression efficiency for most of the mutant tRNAs. However, several exceptions were noted: the A1 tRNA has a low suppression efficiency because of an impaired EF-Tu interaction, but a high aminoacylation efficiency that results in a nonphysiological accumulation of glutaminyl-tRNA; the A31⋅U39 and G27⋅U43 tRNAs increase suppression efficiency via ribosomal performance without altering aminoacylation efficiency; and the C32-A38 tRNA increases both aminoacylation and ribosome performance but enhances the latter function to a greater extent.
CONCLUSION
The results presented here identify mutations at eight base pair positions in disparate regions of tRNA that increase aminoacylation by GlnRS. The three-dimensional structure of the tRNAGln-GlnRS complex shows the enzyme interacting with the phosphate-sugar backbone at positions in tRNAGln corresponding to every mutation in the starting tRNA (Fig. 1 A and C). Thus, these backbone-mediated interactions are important for aminoacylation because other tRNAGln determinants present in the starting tRNA are insufficient to allow aminoacylation on their own. The mutations apparently work by creating wobble base pairs that adjust the local RNA conformation or stability. However, except for the previously described determinant at the end of the acceptor helix of tRNAGln, all of the aminoacylation determinants of tRNAGln described here are without precedence. Nevertheless, we are able to explain every mutation by a specific aspect of the tertiary structure of the complex.
Our results emphasize that synthetase recognition of tRNA is more elaborate than amino acid side chains of the enzyme interacting with nucleotide bases of the tRNA. Recognition also includes synthetase interaction with tRNA backbone functionalities whose distinctive locations in three-dimensional space are exquisitely determined by the nucleotide sequence of the tRNA. The nucleotides whose backbones help determine aminoacylation of tRNAGln are, in fact, among the distinguishing nucleotides of the molecule (24). The prevalence of backbone-mediated interactions in other RNA–protein systems suggests that these interactions generally are important. It is pertinent that an unusual backbone geometry defined by the G⋅U wobble pair in tRNAAla is indicative of this molecule’s aminoacylation by the alanine enzyme (12).
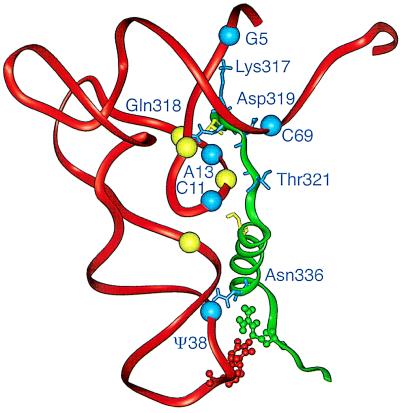
The 2.5-Å structure of the tRNAGln–GlnRS complex (6) reveals that a small protein motif interacts with the phosphate-sugar backbone (but not with the base) of tRNAGln at five mutational sites in the starting tRNA (Fig. 6). The motif also binds the backbone proximal to two other mutational sites (the G15–C48 pair). The motif is extraneous to the catalytic center and has an approximate S shape formed by a strand-alpha-helix-strand (residues Thr-316 to Arg-341). The protein motif binds the backbone at the inside corner of the L-shaped tRNA structure, the flanking acceptor stem, and the anticodon loop of the molecule. The interactions help position the acceptor end of the tRNA in the enzyme’s catalytic site. Table 2 lists these interactions as well as other interactions that were not identified mutationally. Some backbone interactions involve conserved or semiconserved nucleotides such as U8, A14, C11, and C25 that help build the tertiary structure of tRNA (Fig. 1B). The motif also interacts with the base but not the backbone of U35 via Arg-341.
Figure 6.
Molecular structure of part of the tRNAGln–GlnRS complex. The diagram shows the Thr-316–Arg-341 motif of GlnRS and the phosphate-sugar backbone of tRNAGln based on the 2.5-A resolution structure of the complex (6). Amino acid side chains shown are Thr-316, Lys-317, Gln-318, Asp-319, Thr-321, Ser-326, Asn-336, and Arg-341. Spheres representing the phosphate-sugar backbone of residues that interact with the motif are shown (P of residues 5, 8, 11, 12, 13, 14, 25, 38, and 69). Blue spheres indicate nucleotides that are mutationally identified in this work; they interact with amino acid side chains indicated in blue. Arg-341 interacts with the base of U35. Table 2 lists the interacting atoms. The figure was made with insight ii (96.0.6).
The multiple interactions between the amino-terminal strand of the motif and the nucleotide backbone in the core of the tRNA (U8, C11, C12, A13, A14, and C25) could tether the macromolecules and help position the motif’s extremities for other interactions. We note that despite the presence of U35 in the starting tRNA and the functionally subordinate role of Ψ38, a mutation at position 38 nevertheless makes the molecule a better substrate for GlnRS. Enzyme binding changes the conformation of the anticodon region of the tRNA (6), but we do not know whether binding also changes the conformation of the enzyme in general or the motif in particular. The multiple interactions between the motif and the backbone of the tRNA might act as a cooperative unit because any of the mutations increase aminoacylation of the starting tRNA. This all-or-none response would not be obtained with independent interactions.
The same small motif was identified in a genetic analysis of Escherichia coli GlnRS (8). This study relied on the observation that the interaction of Arg-341 with the base of U35 is critical for aminoacylation, and that GlnRS only poorly aminoacylates tRNAGln carrying a C35 substitution. Thus, opal suppressor tRNAGln (UCA anticodon; C35) is inactive, but mutations at either one of two residues in GlnRS partially restore suppressor activity and aminoacylation of tRNAGln. These mutations are Lys-317 to Arg-317 and Gln-318 to either Lys-318 or Agr-318. Because the functional head of Lys-317 interacts with G5 and also with a protein domain that binds the acceptor end of tRNA, a network of interactions functionally connects determinants in the anticodon with those in the acceptor end of the molecule. In summary, mutational analyses of two entirely different systems, one based on the tRNA reported here and the other based on the enzyme (8), have identified the same complementary residues in the macromolecules, G5 paired with Lys-317 and A13 paired with Gln-318. This concurrence is a powerful argument that the residues contribute to aminoacylation.
Another noteworthy feature is that the three-dimensional structures of several other class I aaRS enzymes contain a motif with significant structural similarity to that of GlnRS, but with a different primary sequence. Crystal structures have been determined for 5 of the 10 class I enzymes (1). Comparison of these structures with that of the tRNAGln–GlnRS complex, in combination with tRNA modeling (because the structures lack tRNA), strongly indicates that a small motif similar to that in GlnRS also is present in MetRS (residues 354–378) (25) and GluRS (residues 300–321) (26) enzymes, but is absent in TyrRS and TrpRS enzymes. Other studies indicate that the general orientation of the tRNA substrates on TyrRS and TrpRS is distinct from that in other class I systems (27). Because class I enzymes generally rely on the anticodon and acceptor stem of their substrate tRNAs for aminoacylation, it is possible that the common motif binds the same conserved nucleotides in other tRNAs as in tRNAGln.
Finally, GlnRS and other class I enzymes share a similar active site architecture that binds the acceptor end of the tRNA. In the evolution of protein synthesis, a version of this protein architecture probably was present in smaller synthetases when they interacted with smaller tRNA substrates containing only the acceptor stem sequences corresponding to contemporary tRNAs. The anticodon was in a different location in the smaller tRNAs. As the lengths of synthetases and tRNAs increased, the interaction of the enzyme with the anticodon in its present location was introduced. The motif under discussion may have contributed to this transition by interacting with two critical parts of contemporary tRNAs: backbone functionalities at the junction of the acceptor stem and core region, where the L-shape structure that is common to tRNAs is defined, and a base functionality in the tRNA anticodon, where both amino acid-acceptor and codon specificities of the molecule are defined.
Acknowledgments
We thank L. Silvian and T. A. Steitz for crystallographic information, M. Ibba and D. Söll for GlnRS antibody and marker GlnRS, U. L. RajBhandary for plasmid pAC1, P. Nissen, M. Sprinzl, and M. Nesper for EF-Tu and protocols, and M. Ibba, P. Nissen, W. G. Scott, L. Silvian, D. Söll, M. Sprinzl, P. Strazewski, and M. Yarus for comments on the manuscript. This work was supported by Grant GM42123 from the National Institute of General Medical Sciences, National Institutes of Health.
ABBREVIATIONS
- aaRS
aminoacyl-tRNA synthetase
- GlnRS
glutaminyl-tRNA synthetase
- X-gal
5-bromo-4-chloro-3-indolyl-β-d-galactoside
- EF-Tu
elongation factor Tu
References
- 1.Arnez J G, Moras D. Trends Biochem Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 2.Rould M A, Perona J J, Söll D, Steitz T A. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 3.Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry J C, Moras D. Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 4.Biou V, Yaremchuk A, Tukalo M, Cusack S. Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 5.Goldgur Y, Mosyak L, Reshetnikova L, Ankilova V, Lavrik O, Khodyreva S, Safro M. Structure. 1997;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 6.Rould M A, Perona J J, Steitz T A. Nature (London) 1991;352:213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- 7.Jahn M, Rogers M J, Söll D. Nature (London) 1991;352:258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- 8.Rogers M J, Adachi T, Inokuchi H, Söll D. Proc Natl Acad Sci USA. 1994;91:291–295. doi: 10.1073/pnas.91.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibba M, Hong K-W, Sherman J M, Söll D. Proc Natl Acad Sci USA. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClain W H, Schneider J, Gabriel K. Biochimie. 1993;75:1125–1136. doi: 10.1016/0300-9084(93)90012-h. [DOI] [PubMed] [Google Scholar]
- 11.Westhof E, Dumas P, Moras D. J Mol Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 12.Ramos A, Varani G. Nucleic Acids Res. 1997;25:2083–2090. doi: 10.1093/nar/25.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas R, Wahl M C, Ban C, Sundaralingam M. J Mol Biol. 1997;267:1149–1156. doi: 10.1006/jmbi.1997.0936. [DOI] [PubMed] [Google Scholar]
- 14.Gabriel K, Schneider J, McClain W H. Science. 1996;271:195–197. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 15.McClain W H, Gabriel K, Schneider J. RNA. 1996;2:105–109. [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess R R, Knuth M W. In: Strategies for Protein Purification and Characterization. Marshak D R, Kadonaga J T, Burgess R R, Knuth M W, Brennan W A Jr, Lin S-H, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 227–234. [Google Scholar]
- 17.Smith J D, Celis J E. Nat New Biol. 1973;243:66–71. [PubMed] [Google Scholar]
- 18.Fischer W, Doi T, Ikehara M, Ohtsuka E, Sprinzl M. FEBS Lett. 1985;192:151–154. doi: 10.1016/0014-5793(85)80062-4. [DOI] [PubMed] [Google Scholar]
- 19.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F C, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 20.Arnez J G, Steitz T A. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 21.Yarus M. Science. 1982;218:646–652. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]
- 22.Schultz D, Yarus M. J Mol Biol. 1994;235:1381–1394. doi: 10.1006/jmbi.1994.1095. [DOI] [PubMed] [Google Scholar]
- 23.Yarus M, Smith D. In: tRNA: Structure, Biosynthesis, and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 443–469. [Google Scholar]
- 24.McClain W H. J Mol Biol. 1993;234:257–280. doi: 10.1006/jmbi.1993.1582. [DOI] [PubMed] [Google Scholar]
- 25.Perona J J, Rould M A, Steitz T A, Risler J-L, Zelwer C, Brunie S. Proc Natl Acad Sci USA. 1991;88:2903–2907. doi: 10.1073/pnas.88.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nureki O, Vassylyev D G, Katayanagi K, Shimizu T, Sekine S, Kigawa T, Miyazawa T, Yokoyama S, Morikawa K. Science. 1995;267:1958–1995. doi: 10.1126/science.7701318. [DOI] [PubMed] [Google Scholar]
- 27.Doublié S, Bricogne G, Gilmore C, Carter C W., Jr Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]