Abstract
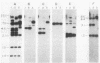
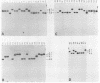
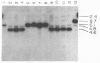
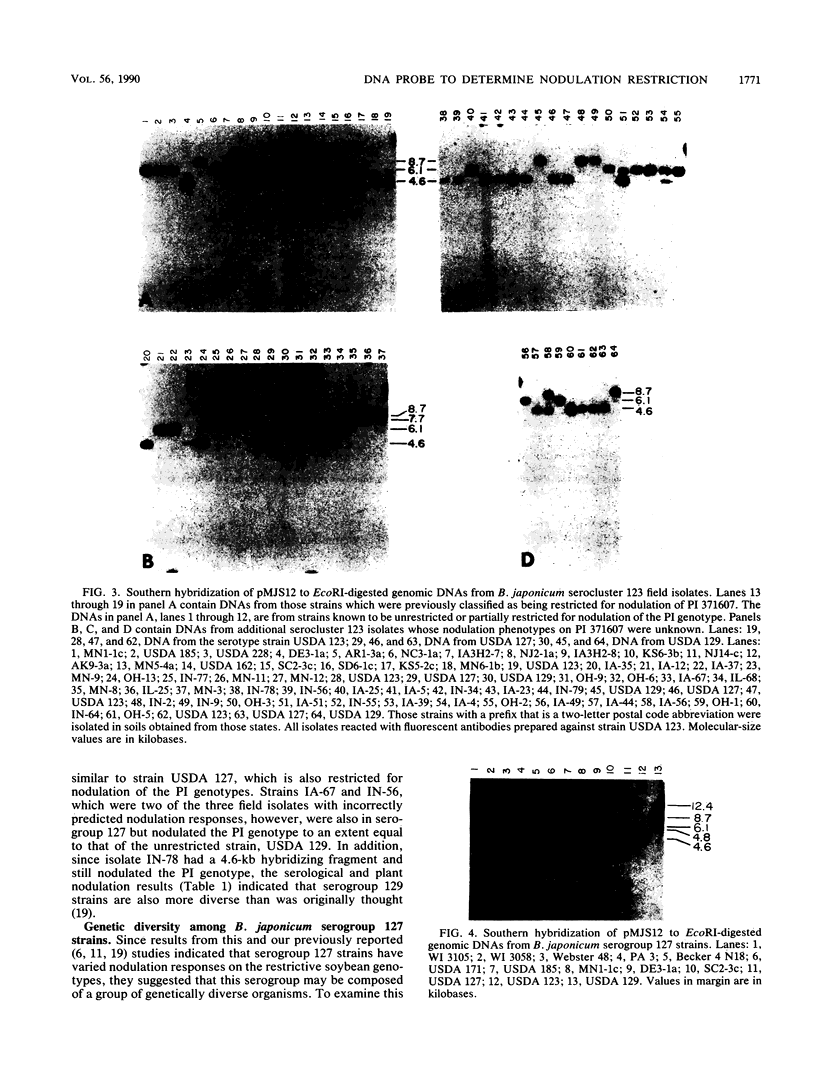
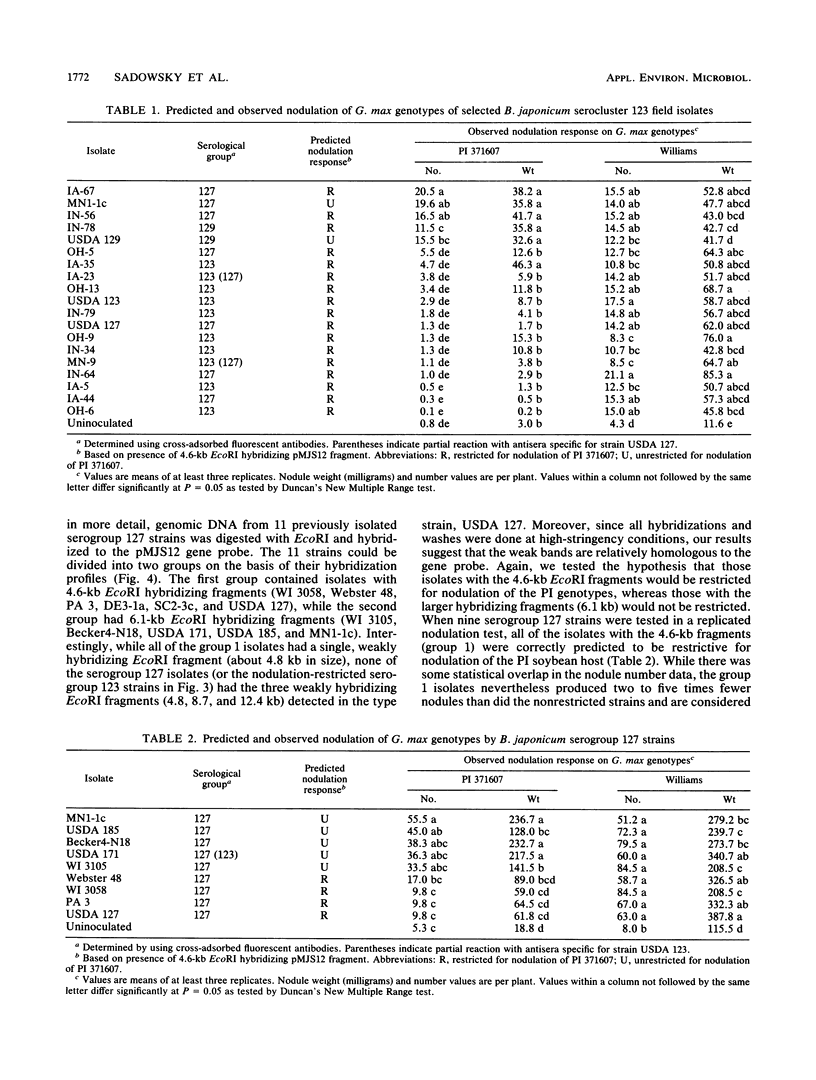
Several soybean plant introduction (PI) genotypes have recently been described which restrict nodulation of Bradyrhizobium japonicum serocluster 123 in an apparently serogroup-specific manner. While PI 371607 restricts nodulation of strains in serogroup 123 and some in serogroup 127, those in serogroup 129 are not restricted. When DNA regions within and around the B. japonicum I-110 common nodulation genes were used as probes to genomic DNA from the serogroup strains USDA 123, USDA 127, and USDA 129, several of the probes differentially hybridized to the nodulation-restricted and -unrestricted strains. One of the gene regions, cloned in plasmid pMJS12, was subsequently shown to hybridize to 4.6-kilobase EcoRI fragments from DNAs from nodulation-restricted strains and to larger fragments in nodulation-unrestricted strains. To determine if the different hybridization patterns could be used to predict nodulation restriction, we hybridized pMJS12 to EcoRI-digested genomic DNAs from uncharacterized serocluster 123 field isolates. Of the 36 strains examined, 15 were found to have single, major, 4.6-kilobase hybridizing EcoRI fragments. When tested for nodulation, 80% (12 of 15) of the strains were correctly predicted to be restricted for nodulation of the PI genotypes. In addition, hybridization patterns obtained with pMJS12 and nodulation phenotypes on PI 371607 indicated that there are at least three types of serogroup 127 strains. Our results suggest that the pMJS12 gene probe may be useful in selecting compatible host-strain combinations and in determining the suitability of field sites for the placement of soybean genotypes containing restrictive nodulation alleles.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayanaba A., Weiland K. D., Zablotowicz R. M. Evaluation of Diverse Antisera, Conjugates, and Support Media for Detecting Bradyrhizobium japonicum by Indirect Enzyme-Linked Immunosorbent Assay. Appl Environ Microbiol. 1986 Nov;52(5):1132–1138. doi: 10.1128/aem.52.5.1132-1138.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfalvi Z., Nieuwkoop A., Schell M., Besl L., Stacey G. Regulation of nod gene expression in Bradyrhizobium japonicum. Mol Gen Genet. 1988 Nov;214(3):420–424. doi: 10.1007/BF00330475. [DOI] [PubMed] [Google Scholar]
- Cooper J. E., Bjourson A. J., Thompson J. K. Identification of lotus rhizobia by direct DNA hybridization of crushed root nodules. Appl Environ Microbiol. 1987 Jul;53(7):1705–1707. doi: 10.1128/aem.53.7.1705-1707.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan P. B., Keyser H. H., Sadowsky M. J. Host Plant Effects on Nodulation and Competitiveness of the Bradyrhizobium japonicum Serotype Strains Constituting Serocluster 123. Appl Environ Microbiol. 1989 Oct;55(10):2532–2536. doi: 10.1128/aem.55.10.2532-2536.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DATE R. A., DECKER A. M. MINIMAL ANTIGENIC CONSTITUTION OF 28 STRAINS OF RHIZOBIUM JAPONICUM. Can J Microbiol. 1965 Feb;11:1–8. doi: 10.1139/m65-001. [DOI] [PubMed] [Google Scholar]
- Göttfert M., Lamb J. W., Gasser R., Semenza J., Hennecke H. Mutational analysis of the Bradyrhizobium japonicum common nod genes and further nod box-linked genomic DNA regions. Mol Gen Genet. 1989 Feb;215(3):407–415. doi: 10.1007/BF00427037. [DOI] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop A. J., Banfalvi Z., Deshmane N., Gerhold D., Schell M. G., Sirotkin K. M., Stacey G. A locus encoding host range is linked to the common nodulation genes of Bradyrhizobium japonicum. J Bacteriol. 1987 Jun;169(6):2631–2638. doi: 10.1128/jb.169.6.2631-2638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Schell M. G., Nelson K. K., Halverson L. J., Sirotkin K. M., Stacey G. Isolation and characterization of the DNA region encoding nodulation functions in Bradyrhizobium japonicum. J Bacteriol. 1985 Dec;164(3):1301–1308. doi: 10.1128/jb.164.3.1301-1308.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Tully R. E., Cregan P. B., Keyser H. H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl Environ Microbiol. 1987 Nov;53(11):2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Bakole R. O., Bohlool B. B. Fluorescent-antibody approach to study of rhizobia in soil. J Bacteriol. 1968 Jun;95(6):1987–1992. doi: 10.1128/jb.95.6.1987-1992.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Zidwick M. J., Abebe H. M. Bradyrhizobium japonicum Serocluster 123 and Diversity among Member Isolates. Appl Environ Microbiol. 1986 Jun;51(6):1212–1215. doi: 10.1128/aem.51.6.1212-1215.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Gibson A. H., Dudman W. F., Watson J. M. Evidence for genetic exchange and recombination of Rhizobium symbiotic plasmids in a soil population. Appl Environ Microbiol. 1987 Dec;53(12):2942–2947. doi: 10.1128/aem.53.12.2942-2947.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatcroft R., Watson R. J. A Positive Strain Identification Method for Rhizobium meliloti. Appl Environ Microbiol. 1988 Feb;54(2):574–576. doi: 10.1128/aem.54.2.574-576.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]