Abstract
Nuclear import of proteins containing a nuclear localization signal (NLS) is dependent on the presence of a cytoplasmic NLS receptor, the GTPase Ran, and p10/NTF2. The NLS receptor is a heterodimeric protein consisting of subunits of approximately 60 and 97 kDa, which have been termed importin α/β, karyopherin α/β, or PTAC 58/97. Members of the 60-kDa/importin α subunit family directly bind to the NLS motif and have been shown to function as adaptors that tether NLS-containing proteins to the p97/importin β subunit and to the downstream transport machinery. Herein we report the identification and characterization of hSRP1γ, a human importin α homologue. The hSRP1γ protein is around 45% identical to the two previously identified human importin α homologues hSRP1α/Rch1 and NPI/hSRP1. hSRP1γ can form a complex with importin β and is able to mediate import of a BSA-NLS substrate in an in vitro nuclear import system. Interestingly, hSRP1γ shows a very selective expression pattern and is most abundantly expressed in skeletal muscle, representing more than 1% of the total protein in this tissue. A potential role for hSRP1γ in tissue-specific transport events is discussed.
After their biosynthesis in the cytoplasm, many nuclear proteins have to be actively transported into the nucleus to reach their functional compartment. To cross the nuclear envelope proteins have to travel through the nuclear pore complex (NPC), a large multiprotein structure that regulates the selective exchange of proteins, RNA, and nucleoprotein complexes between the cytoplasmic and nuclear compartments (1–3). The NPC allows diffusion of molecules smaller than 40–60 kDa in mass but larger proteins and complexes have to be actively transported through the NPC by energy-dependent mechanisms (4). Many nuclear proteins contain a nuclear localization signal (NLS) in their primary amino acid sequence that is recognized by the cellular transport machinery. The NLS is usually characterized by a single or bipartite cluster of basic amino acid residues (boldface type below) exemplified in the NLS of the simian virus 40 (SV40) large tumor antigen (PKKKRKV) or in the NLS of nucleoplasmin (KRPAATKKAGQAKKK) (5). But other NLS variations have also been identified [e.g., the NLSs of c-myc (6) and the NP protein from influenza virus (7)].
The development of in vitro transport systems (8, 9) has allowed the identification and characterization of factors that are necessary to mediate nuclear protein import. The NLS is recognized in the cytoplasm by a heterodimeric NLS receptor that binds to the NPC and mediates the energy-independent docking reaction of the import substrate at the nuclear envelope (for review, see refs. 10 and 11). The NLS receptor consists of an ∼60-kDa subunit and an ∼97-kDa subunit both of which have been identified multiple times in different species. The small subunit that is primarily responsible for the NLS recognition has been named NLS receptor (12), Srp1p (13), importin α (14), hSRP1α/Rch1 (15, 16), hSRP1/NPI (17, 18), karyopherin α (19), m-importin (20), or pendulin/OHO31 (21, 22). The larger subunit binds directly to NPC proteins and has been termed karyopherin β (23), importin β (24), p97 (25), PTAC 97 (26), or Kap95p (27). The α subunit binds to the β subunit via a short amino-terminal domain (28, 29). This amino-terminal interaction domain is sufficient to direct the import of a reporter protein into the nucleus, suggesting that the α subunit functions as a cytoplasmic adaptor that recognizes NLS-containing proteins and tethers them to importin β and to the downstream transport machinery (28, 29).
Two additional soluble factors are needed to transfer the NLS–protein–receptor complex through the NPC, the GTPase Ran (30, 31) and a small homodimeric protein called p10 or NTF2 (32, 33). Like other GTPases, Ran cycles between GTP- and GDP-bound states and this cycle determines the effects of Ran on downstream targets (for review, see ref. 34). Because of the low intrinsic rates of GTP hydrolysis and GDP/GTP exchange, the Ran cycle has to be regulated by guanosine nucleotide exchange factors and by GTPase-activating proteins, respectively. The major Ran guanosine nucleotide exchange factor is the nuclear protein RCC1, whereas the major Ran GTPase-activating protein is cytoplasmic (34). The compartmentalized distribution of these two Ran regulators is likely to establish a concentration gradient of Ran–GTP and Ran–GDP between nucleoplasm and cytoplasm (10, 11). In vitro experiments suggest that the translocation reaction requires Ran in its GDP-bound form on the cytoplasmic side and free GTP as an energy source (35, 36). Ran in its GTP-bound state can form complexes with importin β and NPC proteins such as RanBP2 and Nup2p (37–39), whereas Ran–GDP interacts with p10/NTF2 (40, 41) or with importin β in the presence of RanBP1 (42). In addition, it was demonstrated that Ran–GTP is able to break the trimeric complex of NLS, protein, and receptor (43), a reaction that might be used to terminate the translocation reaction at the nucleoplasmic site of the NPC. Several models for the translocation mechanism through the pore have been proposed but it is still poorly understood how the Ran–GTP/GDP cycle is used to achieve the import of the NLS–protein–receptor complex into the nucleoplasm (11).
Besides the simple or bipartite NLSs for proteins, other karyophilic signals have been described, most notably for the import of U small nuclear ribonucleoproteins (RNPs) and for hetergeneous nuclear RNPs (hnRNPs), such as the hnRNP A1 protein. The karyophilic motif that directs the import of the hnRNP A1 protein is a 38-aa-long motif that has been termed M9 (44). Transportin, a factor that binds to the M9 motif recently was identified and shown to mediate M9 driven import in an in vitro transport system (45). A yeast homologue of transportin, called Kap104p, was independently identified and shown to bind to a class of yeast mRNA binding proteins (46). Both transportin and Kap104 show low homology to importin β but they interact with their import substrates directly without an adaptor such as importin α (45, 46). Several other importin β homologues have now been identified in yeast and metazoans and it has been suggested that they might function as specialized carriers for specific transport events (11, 47, 48).
Herein, we report the cloning and characterization of a human importin α homologue, termed hSRP1γ. hSRP1γ shows a very selective expression pattern and is predominantly expressed in skeletal muscle. It is able to bind to importin β and can mediate NLS-dependent protein import in an in vitro transport assay. hSRP1γ could function as a tissue-specific adaptor protein that mediates import of a subset of NLS-containing proteins.
MATERIALS AND METHODS
Cloning and Recombinant Protein Expression.
The hSRP1γ cDNA was isolated in a two-hybrid screen with a HeLa cDNA library and the nuclear protein p80-coilin as a bait (15). Full-length cDNA clones for hSRP1γ were isolated by 5′ rapid amplification of cDNA ends (RACE) using poly(A)+ RNA from human skeletal muscle. Four clones derived from four different RACE reactions were sequenced in both orientations to assemble the full-length sequence of hSRP1γ. A full-length clone containing the hSRP1γ cDNA (pKW263) was constructed by ligating one of the original two-hybrid clones with a clone derived from the RACE using an unique internal BamHI site. The NcoI–XhoI fragment of pKW263 containing the complete coding sequence of hSRP1γ was subcloned into the vaccinia expression vector pSM56 (49) to allow for expression of a His6-tagged hSRP1γ fusion protein (expression vector pKW318).
The production of recombinant vaccinia virus and the infection of HeLa cells was performed as described by Janknecht et al. (49). Recombinant His6-hSRP1γ protein was purified from 5 liters of infected HeLa cells by using Ni-NTA (where NTA is nitrilo-tri-acetic acid) agarose beads as described (49). As a control for the purification, the same amount of HeLa cells infected with wild-type virus was also lysed and proteins were purified via Ni-NTA agarose beads (mock purification).
All other recombinant protein purifications were performed as described (15, 29).
Affinity Purifications and Northern Blotting.
The coselection experiment was performed by incubating on a rotating wheel ∼2 μg of hSRP1γ in the presence of ∼3 μg of human importin β for 1 hr at 4°C. Ten microliters of Ni-NTA agarose resin (Qiagen, Chatsworth, CA) was added to 100 μl of the mixture and incubated for an additional 1 hr. Beads were washed three times with IPP buffer (150 mM NaCl/20 mM Tris⋅HCl, pH 7.4/5 mM 2-mercaptoethanol), and bound proteins were eluted in 20 μl of 1× SDS loading buffer containing 300 mM imidazole. Five microliters was loaded on the polyacrylamide gel.
Multitissue Northern blots and dot blots (CLONTECH) were analyzed with randomly labeled 32P-labeled probes (50).
Antibody Production, Western Blotting, and Quantitation.
Rabbit antibodies were raised against the keyhole limpet hemocyanin-coupled peptides SEGYTFQVQDGAPFTFNF, QQQYIFQQCEAPMEGFQL, and GTYNFDPTANLQTKEFNF corresponding to the 18 carboxyl-terminal residues of hSRP1α, NPI, and hSRP1γ, respectively. Standard immunization procedures were used.
The concentration of recombinant proteins in the purified fractions was assessed by separating 0.1 μl of each fraction by SDS/PAGE, and the gels were Coomassie-stained, dried, and analyzed by densitometry as described (51). This method gives the concentration of the protein of interest with a 30% accuracy independent of the purity of the fraction (51). Next, 5 μg (hSRP1α, NPI) or 1 μg (hSRP1γ) of Protein Medleys (CLONTECH) from skeletal muscle, testis, brain, and placenta were separated by SDS/PAGE in a gel containing also 100, 30, 10, 3, and 1 ng of recombinant hSRP1α, NPI, or 500, 100, 25, 5, and 1 ng of recombinant hSRP1γ. Relative quantitation was performed as above, linearity was ascertained for the standards, and protein amount in each tissue was determined accordingly.
Import Assays, Image Recording, and Quantitation.
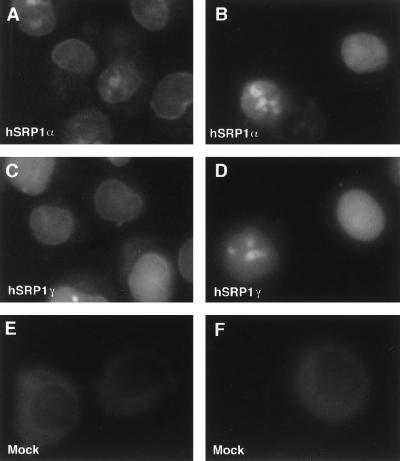
Import assays were performed as described (36). Final concentrations of 0.1 mg/ml for hSRP1α and 0.15 mg/ml for hSRP1γ were used. The equivalent amount of mock-purified extract was added in Fig. 5. Fluorescence samples were observed with a Leica DMLB microscope using a ×100 PL Fluotar oil immersion objective. Images were acquired with an Optronics DEI-750 charge-coupled device camera using the scion image software. Panels were assembled with Adobe photoshop software. Quantitation was performed by scanning a square area containing the whole nucleus and taking the cytoplasmic staining as background by using the gel analyser software (G. Paollela, EMBL).
Figure 5.
In vitro nuclear protein import assays. Import of fluorescein-labeled BSA-NLS conjugate into digitonin-permeabilized HeLa cells was assayed in the presence of hSRP1α and importin β (A); hSRP1α, importin β, Ran, and p10/NTF2 (B); hSRP1γ and importin β (C); hSRP1γ, importin β, Ran, and p10/NTF2 (D); mock-purified extract and importin β (E); or mock-purified extract, importin β, Ran, and p10/NTF2 (F). The localization of the BSA-NLS substrate was analyzed by epifluorescence. Images were recorded by using a charge-coupled device camera. Quantitation of the uptake of BSA-NLS in B and D shows that hSRP1γ has only ∼30% of the import activity of hSRP1α. The protein amounts of hSRP1γ and α used in the assay were normalized with respect to their binding activity to importin β. Coomassie blue staining showed that roughly equal amounts of the two proteins were present in the transport assay (data not shown).
RESULTS
Cloning of hSRP1γ.
We have identified the human NLS-binding protein hSRP1α in a two-hybrid screen with a HeLa cDNA library and the nuclear protein p80-coilin as a bait (15). During this screen 90 positive clones were identified, seven of which coded for hSRP1α. Two additional clones coded for a cDNA that showed high similarity to hSRP1α and to Xenopus importin α on the protein level. Because two other human importin α homologues had been identified, hSRP1α/Rch1 (15, 16) and hSRP1/NPI (17, 18), we decided to term this cDNA hSRP1γ.
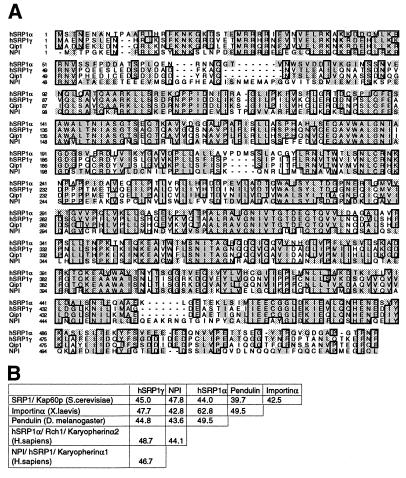
We next wanted to isolate full-length clones for hSRP1γ because neither of the two isolated cDNA clones coded for the complete amino terminus of hSRP1γ. Several attempts to obtain additional 5′ cDNA sequence from different human cDNAs and cDNA libraries failed, a fact that can be explained by the tissue specificity of hSRP1γ’s RNA expression pattern (see below). However, further 5′ cDNA sequence could be obtained by 5′ RACE (52) from skeletal muscle mRNA. The full-length hSRP1γ cDNA is predicted to encode a protein of 58.4 kDa with a calculated isoelectric point of 4.84 (Fig. 1A). Like the other previously identified importin α homologues, hSRP1γ contains eight central “arm repeats” (57) and has an amino-terminal domain that is very rich in basic amino acid residues (Fig. 1A). hSRP1γ shows 48.7% identity on the amino acid level to hSRP1α, 46.7% identity to NPI, and 85.4% identity to the recently identified Qip1 (58) (Fig. 1B). All four human importin-α-like proteins have around 50% identity to the yeast Saccharomyces cerevisiae homologue ySRP1p (13).
Figure 1.
(A) Multiple sequence alignment of different human importin α homologues. The amino acid sequence of hSRP1γ, hSRP1α (GenBank U09559), Qip1 (GenBank AB002533), and NPI/hSRP1 (GenBank S75295) were aligned with clustal w (53) and the alignment was displayed with seqvu using the ges homology matrix. Boxes indicate that at least three out of four amino acids are identical at that position and shading indicates a homology of more than 85%. (B) Identity matrix of hSRP1γ (this study), SRP1 (S. cerevisiae, GenBank M75849), Importin α (Xenopus laevis, GenBank G623602), Pendulin (Drosophila melanogaster, GenBank S57866), hSRP1α (Homo sapiens), and NPI (Homo sapiens). Alignment was performed with fasta (54), and the percentage of identity between the indicated amino is displayed.
To test whether hSRP1γ is able to complement a deletion of the SRP1 gene in S. cerevisiae, a plasmid expressing hSRP1γ was introduced into a yeast strain in which the genomic copy of SRP1 was disrupted (13). The cells were covered by an URA3-marked plasmid containing the yeast SRP1 gene. After plasmid loss by counterselection on 5′-fluoroorotic acid no viable colonies could be recovered (data not shown). Because Western blotting confirmed that hSRP1γ was expressed in the analyzed strain (data not shown), it can be concluded that hSRP1γ is not able to rescue an SRP1 deletion in S. cerevisiae. Furthermore, neither hSRP1α or NPI alone nor double or triple combinations of the three analyzed importin α homologues were able to substitute for the loss of SRP1 in S. cerevisiae (data not shown).
Tissue Specificity of hSRP1γ Expression.
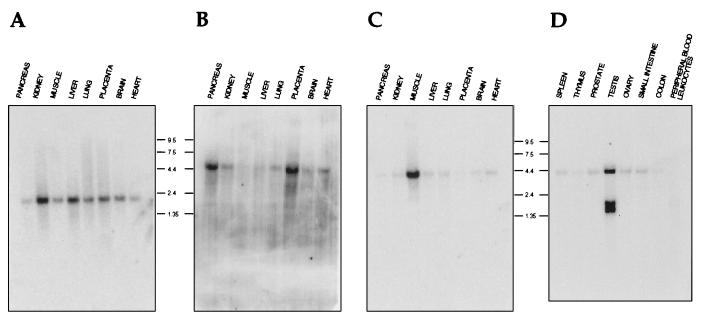
The observation that the cDNA encoding hSRP1γ was not present in several cDNA libraries from different tissues prompted us to analyze the RNA expression pattern of hSRP1γ (Fig. 2). First, a human multitissue Northern blot was analyzed with hSRP1α (Fig. 2A), NPI (Fig. 2B), and hSRP1γ (Fig. 2C) cDNA probes. A 2,200-nt RNA hybridizing to the hSRP1α probe could be detected in all tissues tested albeit with slightly lower levels in pancreas, skeletal muscle, and heart muscle (Fig. 2A). The NPI expression, revealed by a hybridizing transcript of around 4,300 nucleotides, was more variable showing the highest levels in pancreas, placenta, and heart (Fig. 2B). In contrast, expression of a 4,000-nt-long mRNA of hSRP1γ could be mainly detected in skeletal muscle, and only minor amounts of transcripts were recognized in mRNAs isolated from pancreas, kidney, liver, lung, placenta, brain, or heart (Fig. 2C). An additional human multitissue Northern blot was analyzed with an hSRP1γ probe to further characterize hSRP1γ’s RNA expression pattern (Fig. 2D). High levels of three transcripts of approximately 4,000, 1,500, and 1,400 nucleotides could be detected in testis, but basically no transcripts were present in spleen, thymus, prostate, ovary, small intestine, or colon. It is presently unclear whether the smaller transcripts are functional mRNAs because the ORF for hSRP1γ consists of 1,563 nucleotides. Dot-blot analysis did not reveal significant expression of hSRP1γ in different subregions of the brain (cerebral cortex, amygdala, cerebellum, hippocampus, and spinal cord) or in bladder, aorta, uterus, stomach, lymph node, or bone marrow (data not shown).
Figure 2.
Northern blots showing the expression pattern of hSRP1α (A), NPI/hSRP1 (B), and hSRP1γ (C and D) and in different human tissues. Two multitissue Northern blots containing poly(A)+ RNA isolated from the indicated human tissues were analyzed with probes derived from the cDNAs of hSRP1α (A), NPI (B), or hSRP1γ (C and D). Molecular sizes are in kilobases.
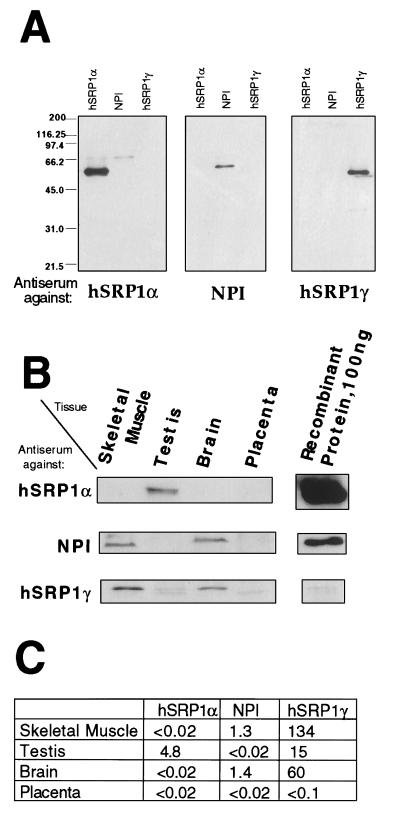
To analyze the relevance of the RNA expression pattern, we also studied the protein expression in human skeletal muscle, testis, brain, and placenta, by using specific antibodies raised against three importin α homologues (Fig. 3). Surprisingly, the expression patterns observed by Western blotting displayed some marked differences when compared with the mRNA expression. For instance, a significant amount of hSRP1γ protein could be detected in brain where hardly any hSRP1γ transcript could be found. This suggests a posttranscriptional regulation of hSRP1γ’s expression in brain. However, the very high levels of expression of hSRP1γ in muscle were confirmed at the protein level. In this tissue, the hSRP1γ was estimated to represent between 0.9 and 1.7% of the total protein mass. Thus, in skeletal muscle hSRP1γ is at least 100 times more abundant than NPI, the only other homologue detected in that tissue.
Figure 3.
Protein expression pattern of hSRP1γ, hSRP1α, and NPI/hSRP1 in different human tissues. (A) Specificity of the antisera used in this study. About 500 ng of the indicated recombinant proteins were separated by SDS/PAGE, transferred to nitrocellulose, and subsequently probed with the antiserum against hSRP1α, NPI, or hSRP1γ. (B) Multitissue Western blots probed with the antiserum against hSRP1α, NPI, or hSRP1γ. Five micrograms (hSRP1α, NPI) or 1 μg (hSRP1γ) of tissue protein extracts (CLONTECH) and 50 μg (hSRP1α, NPI) or 10 μg (hSRP1γ) were loaded in the first four lanes. The indicated amount of recombinant protein loaded in the last lane was determined by densitometry scanning in a Coomassie-stained gel. (C) Result of quantitation of protein expression. The values correspond to the amount (in ng) of each importin-α-like factor per 10 μg of total protein.
hSRP1γ Can Form a Complex with Importin β.
Because hSRP1γ has only around 50% identity to hSRP1α and NPI, it was necessary to analyze whether hSRP1γ also could function as a cytoplasmic adaptor that mediates interactions of NLS-containing proteins with importin β and the downstream components of the transport machinery.
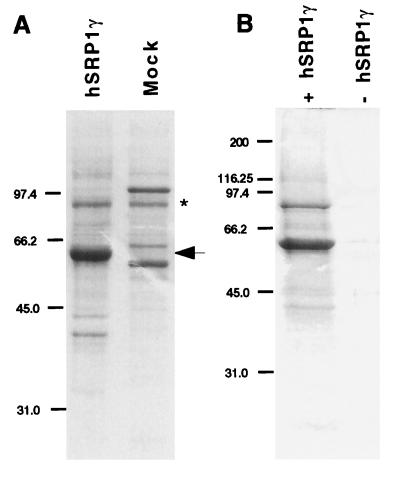
hSRP1γ was expressed in the vaccinia virus expression system (49) because we were unable to express active protein in Escherichia coli. His6-hSRP1γ was purified from infected HeLa cells and its purity was analyzed by Coomassie staining after separation by SDS/PAGE (Fig. 4A). A faint band of ∼90 kDa copurified with hSRP1γ (Fig. 4A, lane 1) but an identical band also was detected after mock purification from cells infected with wild-type virus (Fig. 4A, lane 2). Western blotting with an antibody specific for importin β demonstrated that this ∼90-kDa band is not importin β (data not shown).
Figure 4.
(A) Expression and purification of recombinant His6-hSRP1γ. Histidine-tagged hSRP1γ was expressed in HeLa cells with the vaccinia virus expression system and purified over Ni-NTA agarose, and its purity was analyzed by Coomassie staining after separation on 10% SDS/PAGE gels (lane 1, hSRP1γ). A similar purification was performed from HeLa cells that had been infected with the wild-type vaccinia virus (lane 2, mock). The arrow indicates hSRP1γ. The asterisk shows a contaminating protein of ∼90 kDa that also was purified from the wild-type-infected extract. Western blotting demonstrated that this band is not importin β (data not shown). Molecular sizes are in kilodaltons. (B) Coselection of importin β by hSRP1γ. Untagged recombinant importin β was incubated with Ni-NTA agarose beads either in the presence (lane 1) or absence (lane 2) of His6-tagged hSRP1 γ. After selection the proteins were separated on a 10% SDS/PAGE gel and analyzed by Coomassie staining.
To test whether hSRP1γ can interact with importin β, coselection experiments on Ni-NTA beads were performed with recombinant importin β, either in the presence (Fig. 4B, lane 1) or absence of His6-tagged hSRP1γ (Fig. 4B, lane 2), and the selected proteins were analyzed by Coomassie staining after separation on 10% SDS/PAGE gels. Importin β was specifically coselected only in the presence of hSRP1γ, demonstrating that hSRP1γ can form a stable complex with importin β in vitro. Similar experiments using [35S]-labeled importin β further confirmed a direct interaction of these two proteins in vitro (data not shown).
hSRP1γ Functions as a Nuclear Transport Factor.
Next, we wanted to test whether hSRP1γ is able to mediate nuclear import of a BSA-SV40 NLS reporter in an in vitro import system (Fig. 5). As reported (29), the recombinant human transport factors hSRP1α and p97 alone mediate the docking of a BSA-NLS reporter substrate at the NPC (Fig. 5A). Addition of Ran–GDP, p10, and energy resulted in efficient nuclear uptake of the BSA-NLS reporter (Fig. 5B). When hSRP1γ was used, instead of hSRP1α, again accumulation of BSA-NLS at the nuclear envelope was observed in the presence of importin β (Fig. 5C). Complete nuclear import was achieved when hSRP1γ, importin β, Ran, p10, and energy were present (Fig. 5D). Both the docking and the import reaction were strictly dependent on the presence of an importin-α-like factor because neither docking nor translocation occurred in the presence of the mock-purified extract and importin β alone (Fig. 5E) or with importin β, Ran-GDP, p10, and energy (Fig. 5F).
It should be noted that nuclear import of the BSA-NLS substrate was consistently lower with hSRP1γ than with hSRP1α when similar amounts of active hSRP1α and hSRP1γ were present in the assays (importin β binding activity was used as a tool to estimated protein activity). Quantitation of the fluorescence signals shows that hSRP1α has a roughly 3-fold higher import activity than hSRP1γ (data not shown). Because identical amounts of importin β were used in the import assays, the most likely interpretation is a difference in the relative affinity of these two factors toward the SV40 NLS motif. In any event, it can be concluded that hSRP1γ functions as a nuclear transport factor and that it is a genuine analogue of importin α. These data show that hSRP1γ is able to bind to a BSA-NLS reporter substrate in the presence of importin β and that it can mediate the interaction of its cargo with importin β and other components of the nuclear transport machinery.
DISCUSSION
Nuclear protein import of NLS-containing proteins is mediated by a cytoplasmic NLS receptor complex consisting of two subunits of about 60 and 97 kDa termed importin α/β. Herein, we describe the cloning and characterization of a human importin α homologue, hSRP1γ. hSRP1γ shows around 45% identity to the other two human family members hSRP1α and NPI and around 85% identity to Qip1 (58). Like the other importin α homologues, hSRP1γ is an acidic protein with a calculated isoelectric point of 4.84. It is characterized by a core of eight hydrophobic repeats, each 43 amino acids long, the so-called arm repeats (57). We show herein that hSRP1γ together with importin β is able to bind to a BSA-NLS substrate and mediate its docking at the nuclear envelope. In the presence of Ran and p10/NTF2, the dimeric complex of hSRP1γ and importin β supports complete nuclear import. hSRP1γ is thus a nuclear transport factor that functions in an in vitro assay similar to Xenopus importin α, hSRP1α, and NPI. It should be pointed out that the in vitro import assay used in this study might not necessarily reveal small differences in the function of these three adaptor molecules. For example, the BSA substrate used possesses multiple NLS peptides and functions very efficiently as an in vitro import substrate. Thus affinity differences between the NLS adaptors toward single intrinsic NLS motifs in in vivo substrates cannot be tested with this assay. However, we observed a consistently lower in vitro import activity of hSRP1γ when compared with the activities of hSRP1α or NPI (Fig. 5 and data not shown). The most likely explanation for that behavior is a difference in the affinity of the tested adaptor molecules toward the SV40 NLS, but we cannot entirely exclude that the lower import activity of hSRP1γ is caused by inhibitory factors present in the purified fraction because the recombinant proteins were purified from different sources and with different protocols. However, the proteins have a comparable binding activity when tested in importin β interaction assays (data not shown).
At its amino terminus, hSRP1γ possesses a highly conserved stretch of positively charged amino acids. It has been shown for hSRP1α, NPI (29), and Xenopus importin α (28) that this domain is responsible for the interaction with importin β. Because this amino-terminal domain is highly conserved between hSRP1γ and the other family members and shows an even higher degree of identity than the rest of the protein (e.g., 58% between hSRP1γ and Xenopus importin α, Fig. 1), it is very likely that this amino-terminal domain of hSRP1γ also mediates the direct interaction with importin β.
Database searches revealed several expressed sequence tags coding for additional human importin α homologues (data not shown), indicating the existence of an even larger multigene family of human importin α proteins. This is in contrast to S. cerevisiae where just a single importin α gene (SRP1) exists, but multiple homologues for importin β can be readily identified (refs. 11, 47, and 48 and unpublished data). One importin β homologue, transportin (45), and its yeast orthologue, Kap104p (46), can interact with mRNA binding proteins, and transportin has been shown to mediate the import of the hnRNP A1 protein in an in vitro system (45). Transportin/Kap104p seems to bind directly to its cargo without the need of an adaptor-like protein, such as importin α. It was proposed that different importin β homologues have evolved to bind to different cargoes that need to be transported across the nuclear envelope (11, 48). It also has been suggested that importin β is the archetypical carrier for the classical NLS-dependent protein import and that importin-α-like adaptor proteins have evolved to allow the recognition of different NLS-containing proteins and facilitate their transport by using a common mechanism (29). This is supported by the fact that the amino-terminal importin β binding domain of importin α shows sequence similarity to NLS motifs and functions as a karyophilic signal when conferred to a reporter protein (28, 29). Obviously, the use of a family of adaptors would allow a greater flexibility in the recognition of divergent NLS motifs in different classes of NLS containing proteins. This model is supported by data indicating that hSRP1α and NPI bind with variable specificities to a subset of nuclear proteins (7, 57). The presented data are consistent with this model because hSRP1γ has clearly a lower import activity than hSRP1α in an in vitro import assay using the BSA-SV40 NLS reporter. This model should now be tested in more detail to compare the distinct and/or overlapping NLS-binding characteristics of all known human importin α homologues.
Another intriguing reason for the evolution of an importin α multigene family might be found in the different expression patterns of the three homologues presented herein. As discussed above the genome of S. cerevisiae contains a single gene, SRP1, encoding the yeast importin α protein (13). However, the primitive multicellular organism, the worm Caenorhabditis elegans, contains at least two genes coding for importin α homologues (data not shown). Also Drosophila and Xenopus seem to have two or more genes coding for importin-α-like proteins. In this context it is intriguing that the three human importin α homologues studied herein show selective expression in different tissues both on the RNA and protein level. For example, hSRP1γ is an extremely abundant protein in skeletal muscle and represents probably more than 1% of the total protein in that tissue. hSRP1γ therefore is expressed at least 100 times more than NPI or hSRP1α, which are barely or not at all detectable in skeletal muscle (Fig. 3C). It can be speculated that in metazoans different NLS-binding adaptors have evolved to fulfill tissue-specific functions and they would, therefore, allow a regulation of nuclear protein import in a tissue-specific fashion. This idea might be supported by the fact that one Drosophila family member pendulin/oho31 is not essential for viability and its mutational inactivation causes a malignant transformation of hematopoietic precursor cells (21, 22).
In this regard it is also interesting that neither the three tested importin-α-like factors alone nor all three human factors together were able to rescue a yeast importin α gene (SRP1) deletion (data not shown). This suggests a functional difference between the human and yeast NLS-binding factors especially because it was demonstrated that the SV40 NLS functions in yeast (58) and that the hSRP1α importin-β-binding domain is able to mediate nuclear import of a green fluorescent protein reporter protein (M.V.N. and K.W., unpublished observation). The latter observation indicates that the interaction between the importin α and β subunits is conserved between yeast and human and the most likely explanation for the noncomplementation is, therefore, a difference in either the NLS recognition event or in the recycling of importin-α-like factors from the nucleus to the cytoplasm. The fact that these factors are not simply interchangeable is consistent with the hypotheses that the invention of multiple importin-α-like factors in metazoans reflects a functional diversity and that the individual factors might play a role in different, probably tissue-specific, transport events.
Acknowledgments
We are grateful to V. Sonntag-Buck, H. Holz, and Henk Stunnenberg for their help and advice with the vaccinia virus expression system. We thank M. Nomura for providing the SRP1 deletion strain and P. Palese for the NPI cDNA. We are also grateful to C. S. Ford for her comments on the manuscript and to the colleagues from the P. Walter lab for helpful discussions. M.N. is a fellow of Ecole Normale Supérieure (Paris). K.W. and A.I.L. acknowledge support from the Deutsche Forschungsgemeinschaft and the Wellcome Trust (U.K.), respectively.
ABBREVIATIONS
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- SV40
simian virus 40
- RNP
ribonucleoprotein
- hnRNP
heterogeneous nuclear RNP
- RACE
rapid amplification of cDNA ends
- NTA
nitrilo-tri-acetic acid
Footnotes
Data deposition: The nucleotide sequence reported in this paper has been deposited in the GenBank database (accession no. AF034756).
References
- 1.Davis L I. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 2.Doye V, Hurt E. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 3.Pante N, Aebi U. J Cell Sci (Suppl) 1995;19:1–11. doi: 10.1242/jcs.1995.supplement_19.1. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg M W, Allen T D. Curr Biol. 1995;7:301–309. doi: 10.1016/0955-0674(95)80083-2. [DOI] [PubMed] [Google Scholar]
- 5.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 6.Makkerh J P S, Dingwall C, Laskey R A. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Palese P, O’Neill R E. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newmeyer D D, Finlay D R, Forbes D J. J Cell Biol. 1986;103:2091–2102. doi: 10.1083/jcb.103.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adam S A, Marr R S, Gerace L. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 11.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 12.Adam S A, Gerace L. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 13.Yano R, Oakes M, Yamaghishi M, Dodd J A, Nomura M. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görlich D, Prehn S, Laskey R A, Hartmann E. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 15.Weis K, Mattaj I W, Lamond A I. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 16.Cuomo C A, Kirch S A, Gyuris J, Brent R, Oettinger M A. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortes P, Ye Z S, Baltimore D. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill R E, Palese P. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 19.Moroianu J, Hijikata M, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Küssel P, Frasch M. J Cell Biol. 1995;129:1491–1507. doi: 10.1083/jcb.129.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Török I, Strand D, Schmitt R, Tick G, Török T, Kiss I, Mechler B M. J Cell Biol. 1995;129:1473–1489. doi: 10.1083/jcb.129.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radu A, Blobel G, Moore M S. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Görlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 25.Chi N C, Adam E J H, Adam S A. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamoto N, Shimamoto T, Kose S, Takao T, Tachibana T, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. FEBS Lett. 1995;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- 27.Enenkel C, Blobel G, Rexach M. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 28.Görlich D, Henklein P, Laskey R A, Hartmann E. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 29.Weis K, Ryder U, Lamond A I. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 30.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 31.Melchior F, Paschal B, Evans J, Gerace L. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore M S, Blobel G. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paschal B M, Gerace L. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rush M G, Drivas G, D’Eustachio P. BioEssays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- 35.Görlich D, Pante N, Kutay U, Aebi U, Bischoff F R. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 36.Weis K, Dingwall C, Lamond A I. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama N, Hayashi N, Seki N, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, Fukui M, Nishimoto T. Nature (London) 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 39.Dingwall C, Kandels-Lewis S, Seraphin B. Proc Natl Acad Sci USA. 1995;92:7525–7529. doi: 10.1073/pnas.92.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nehrbass U, Blobel G. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- 41.Clarkson W D, Kent H M, Stewart M. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- 42.Chi N C, Adam E J, Visser G D, Adam S A. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rexach M, Blobel G. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 44.Siomi H, Dreyfuss G. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 46.Aitchison J D, Blobel G, Rout M P. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 47.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rout M P, Blobel G, Aitchison J D. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 49.Janknecht R, de Martynoff G, Lou J, Hipskind R A, Nordheim A, Stunnenberg H G. Proc Natl Acad Sci USA. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 51.Field C M, al-Awar O, Rosenblatt J, Wong M L, Alberts B, Mitchison T J. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dieffenbach C W, Dveksler G S. PCR Primer: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 53.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeifer M, Berg S, Reynolds A B. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 56.Seki T, Tada S, Katada T, Enomoto T. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- 57.Nadler S G, Tritschler D, Haffar O K, Blake J, Bruce A G, Cleaveland J S. J Biol Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- 58.Nelson M, Silver P. Mol Cell Biol. 1989;9:384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]