Abstract
In the yeast Saccharomyces cerevisiae, meiotic recombination is initiated by DNA double-stranded breaks (DSBs) occurring in micrococcal nuclease (MNase)-hypersensitive regions of the chromatin. MNase-sensitive sites also undergo meiosis-specific alterations in chromatin structure prior to the appearance of DSBs. DSB formation requires the products of numerous genes. Herein we have examined the effects of mutations in four such genes, MRE11, RAD50, XRS2, and MRE2, on MNase sensitivity at DSB sites in premeiotic and meiotic cells. Disruption mutations in each of four genes confer greater than wild-type levels of MNase sensitivity in premeiotic cells. In meiotic prophase, all of these mutations affect MNase sensitivity at DSB sites and fall into two distinct phenotypic classes. The type 1 mutations (mre2 and mre11) confer a reduction in MNase sensitivity relative to the wild-type level. The type 2 mutations (rad50 and xrs2) permit a meiotic increase in the MNase sensitivity to reach a final level higher than that observed in wild-type cells. An mre11 disruption mutation (type 1) is epistatic to a rad50 null mutation (type 2) with respect to its meiotic effects on MNase sensitivity, suggesting that the events observed in the type 2 mutants during meiosis are dependent upon type 1 functions. One interpretation of these results is that Mre11, Rad50, Xrs2, and possibly Mer2 (whose splicing is Mre2-dependent) form a complex at recombination hot spots and establish a chromatin/DNA configuration favorable for the induction of DSBs.
Genetic recombination in eukaryotic organisms occurs at a frequency several orders of magnitude higher in meiotic cells than in somatic cells. In the yeast Saccharomyces cerevisiae, most meiotic recombination is initiated at defined sites by the formation of DNA double-stranded breaks (DSBs) that are subsequently repaired by recombination that occurs primarily between homologs (1–11).
The induction of meiotic DSBs is affected by several factors (for reviews, see refs. 12–15). First, DSB formation is controlled by numerous genes including MER2, MRE2, MRE11, RAD50, SPO11, and XRS2 (3, 16–28). In the corresponding null mutants, meiotic DSB formation is absent. Spo11 very likely functions as the catalytic subunit in the meiotic DNA cleavage reaction (29, 30). Mre2 (19) is required for the meiosis-specific splicing of the MER2 transcript (20). Rad50 and Xrs2 have been found to interact with Mre11 (21, 31), and it has been hypothesized that these three proteins required for mitotic DSBs repair form a recombination-initiating complex in meiosis that is essential for the pairing of homologs, DSB formation, and repair (32, 33). The Rad50 and Mre11 proteins are homologous to the Escherichia coli SbcC and SbcD nucleases, respectively (34, 35), which suggests that this complex may have nuclease activity. Furthermore, Rad50 is required for normal development of chromosome structure in meiosis (24) and also for telomere maintenance during mitosis (36).
Second, the formation of DSBs requires an appropriate chromatin substrate. In regions in which chromatin is especially accessible (10, 37, 38) and other requirements are met (11), DSBs occur without apparent DNA sequence specificity (39–41). During meiotic prophase, sensitivity to micrococcal nuclease (MNase) increases locally at a DSB hot spot before the appearance of DSBs (37). Similar chromatin features are observed at the ade6M26 recombination hot spot during meiosis in the yeast Schizosaccharomyces pombe (42). On the other hand, all nuclease-hypersensitive sites are not necessarily DSB hot spots (37, 38, 43). The level of DSBs at nuclease-hypersensitive sites is also influenced by competition between distant hot spots (43–45).
Third, the frequency of DSBs depends on interhomolog interactions (44, 46, 47). In yeast premeiotic cells, homologs are paired via multiple interstitial interactions (48). These interactions disappear during meiotic S phase and are reestablished early in meiotic prophase. The presence of nucleotide sequence heterology in DSB regions causes a reduction in DNase I hypersensitivity during mitosis (49) and a decrease in the frequency of meiotic DSBs (44, 46), suggesting a link between the pathway for DSB formation and the recognition of DNA identity. It is therefore postulated that a recombination complex assembled at nucleosome-free regions in chromatin prior to meiotic DSB formation mediates interhomolog interactions, the recognition of DNA identity, and the formation of DSBs (44, 46, 49).
In the present study, we have analyzed the effects of mre11, rad50, xrs2, and mre2 mutations on MNase sensitivity at DSB sites in premeiotic and meiotic cells. We report that functions provided by the corresponding wild-type genes are required to establish a normal chromatin/DNA configuration at these sites, not only in meiosis but also in premeiosis.
MATERIALS AND METHODS
Yeast Strains.
All strains have the SK1 background. TNY042 is a derivative of NKY1038/NKY1040 (a/a, ho∷LYS2/ho∷LYS2, lys2/lys2, ura3/ura3, leu2∷hisG/ leu2∷hisG, his4X∷LEU2/his4B∷LEU2, arg4-nsp/arg4-bgl, cyh2/CYH2, provided by Nancy Kleckner, Harvard University). TNY047 is isogenic to TNY042 except for mre2∷hisG/mre2∷hisG. Strains AHY104 (mre11∷ URA3/mre11∷URA3), RKD102 (mre11∷hisG-URA3-hisG/ mre11∷hisG-URA3-hisG), XDU278 (xrs2∷URA3/xrs2∷ URA3), KJC210 (rad50∷hisG/rad50∷hisG), KJC312 (mre11∷hisG-URA3-hisG/mre11∷hisG-URA3-hisG, rad50∷ hisG/rad50∷hisG) are isogenic to NKY278 (a/a, ho∷ LYS2/ho∷LYS2, lys2/lys2, ura3/ura3) (24). Disruptants for mre2, mre11, and xrs2 are deletion mutants that retain the N terminus portion and were constructed by insertions of his G, hisG-URA3-hisG (20), and URA3, respectively (21). The rad50∷hisG mutation is a RAD50 null mutation (24).
Presporulation and Sporulation Cultures.
Presporulation and sporulation cultures were as described (50, 51). Briefly, a single colony from a YPG (3% glycerol/2% Bacto Peptone/1% yeast extract) plate was inoculated into 10 ml of SPS presporulation medium (0.5% yeast extract/1% Bacto Peptone/0.17% yeast nitrogen base without ammonium sulfate and amino acids/0.05 M potassium phthalate/1% potassium acetate/0.5% ammonium sulfate, pH 5.0) with nutritional supplements, and cells were cultured at 30°C overnight. For practical reasons, we could examine at most four strains in parallel on the same day. Thus, for the precise comparison of data obtained on different days, we systematically included a control culture of the wild-type strain TNY042 or NKY278. Small amounts of the preculture suspension were then inoculated into 0.5 liter of SPS with supplements, and cells were cultured at 30°C to a density of 2 to 4 × 107 cells per ml. Cells were harvested by centrifugation and washed once in sterile water, and half of the cells were pelleted and frozen in liquid nitrogen as t = 0-h samples. The other half was inoculated into 0.5 liter of SPM (1% potassium acetate/0.001% polypropylene glycol 2,000 in 5-liter flasks) with supplements, and cells were cultured at 30°C for 2, 4, and 6 h with vigorous aeration. To verify that mutant strains undergo meiosis with kinetics similar to those of wild type, the progression of meiosis I in wild-type (TNY042 and NKY278) and mutant strains was followed by 4′,6-diamidino-2-phenylindole staining.
Chromatin Preparation, Digestion of Chromatin by MNase, Hybridization, and Quantification.
Preparation of crude chromatin fractions from S. cerevisiae cells and treatment of chromatin with MNase (7, 10, 20, and 50 units/ml) were performed as described (52). We found that it is essential to avoid prolonged incubation of the cells with zymolyase during spheroplast formation because excess treatment causes partial cell lysis, nucleosomal rearrangements, and a substantial decrease in meiotically induced MNase sensitivity at DSB sites. To minimize experimental variation due to the difference in both the extent of zymolyase treatment and the final concentration of chromatin, samples of chromatin were prepared from a fixed amount of cells [1–2 g (wet weight)]. Indirect end-labeling was performed as described (37). MNase-treated or untreated DNA (7–14 nmol) was digested to completion by PstI. The digested DNA was ethanol-precipitated and separated by electrophoresis on a 1.2% (for ARG4) or 1.5% (for CYS3) agarose gel. Transfer, hybridization, and quantification of band intensities were as described (37). Band intensity was expressed as a percentage of the total band intensity in the lane, including the unbroken parental fragment.
Quality Control of Chromatin Preparations.
Digestion of chromatin with MNase is affected by several factors. Therefore, to accurately compare data for MNase hypersensitivity, we established four criteria for quality control of our chromatin preparations. An experiment was discontinued under any of the following circumstances: (i) the wild-type control strain (TNY042 or NKY278) exhibited an aberrant timing of meiosis, as judged by the formation of DSBs or by the frequency of Arg+ recombinants in return-to-growth conditions; (ii) the ratio of meiotic to premeiotic MNase sensitivities at the ARG4 DSB sites in the wild-type control was lower than 1.5; (iii) the extent of digestion of the CYS3 site I after treatment with 10 units of MNase was higher than 20%, indicating substantial cell lysis during the spheroplast formation; or (iv) a greater than 50% difference between the premeiotic and meiotic MNase sensitivities of a non-DSB site was observed, suggesting that digestion was not properly controlled.
RESULTS
Positioning of MNase-Hypersensitive Sites Is Independent of MRE11, RAD50, XRS2, and MRE2 Functions.
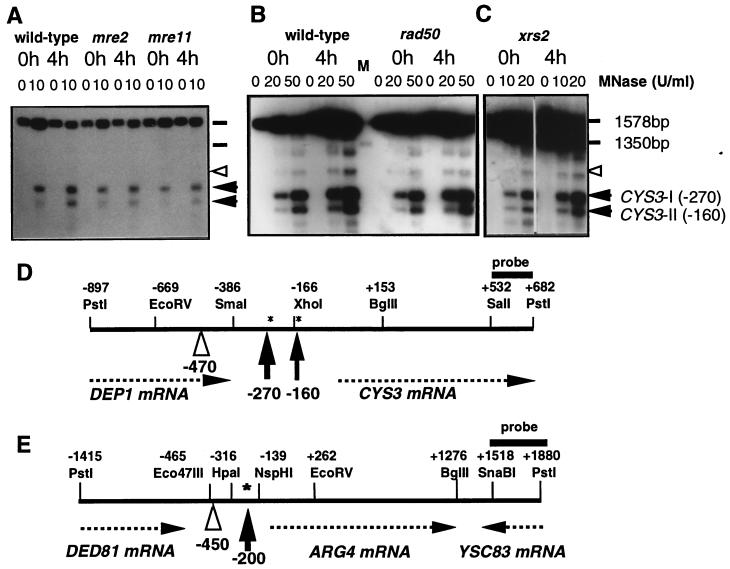
To study the roles of the MRE11, RAD50, XRS2, and MRE2 gene products on the accessibility of DNA in chromatin at meiotic DSB hot spots, we analyzed the effects of mutations in these genes on MNase sensitivity at two DSB hot spots (ARG4 and CYS3) in premeiotic and meiotic cells. Importantly, we systematically included a wild-type diploid in each set of experiments, thereby allowing us to directly compare the behavior of mutant strains to that of wild-type strains. In addition, we applied rigorous criteria to control the quality of the chromatin preparations and to allow precise quantitative comparisons (see below). Isolated chromatin that fulfilled these criteria was treated with various concentrations of MNase, and hypersensitive sites were revealed by the indirect end-labeling method by using probes for the ARG4 and CYS3 loci (37). The data shown in Fig. 1 illustrate typical examples of the detection of MNase-hypersensitive sites in the CYS3 region. At both loci, we found a similar distribution of MNase-sensitive sites in the mutant and wild-type cells in both the premeiotic (t = 0 h) and meiotic (t = 4 h) conditions. This indicates that the absence of the Mre11, Rad50, Xrs2, or Mre2 proteins does not profoundly affect the positioning of accessible regions in chromatin.
Figure 1.
(A–C) Changes in MNase hypersensitivity at the CYS3 hot spot in wild-type and DSB-deficient mutant strains (A, mre2∷URA3, mre11∷URA3; B, rad50Δ; C, xrs2∷URA3). DNA in chromatin from cells cultured in SPM for 0 h (lanes 0h) and 4 h (lanes 4h) was treated with MNase at 0, 10, 20 (0, 20, and 50 for rad50Δ) units/ml. PstI-digested DNA was separated on a 1.5% agarose gel and examined by Southern blot hybridization using a CYS3 PstI–SalI probe. The positions of the two hypersensitive sites at the CYS3 hot spot are shown by arrowheads. A site serving as an internal standard (a noninducible and non-DSB site) is indicated by an open arrowhead. Molecular size standards are indicated with horizontal bars. (D and E) Diagrams of the CYS3 (D) and ARG4 (E) loci. Positions for restriction sites, MNase-hypersensitive sites (shown by vertical arrows), and control MNase-sensitive sites (open arrowheads) for internal standards are indicated relative to position +1, the first base of the CYS3 or ARG4 coding region. Asterisks show the positions of meiotic DSBs. Horizontal arrows represent the orientation of the transcripts indicated. Probes used in the present study are shown by horizontal thick bars.
Mutations in MRE11, RAD50, XRS2, and MRE2 Increase the MNase Sensitivity of DSB Sites in Premeiotic Cells.
For quantitative analysis, we measured the intensity of the bands corresponding to MNase-sensitive signals at DSB sites and at an adjacent control site that is MNase-sensitive but not a DSB hot spot. We calculated two ratios: the ratio of mutant to wild-type MNase sensitivity and the ratio of premeiotic to meiotic sensitivity in a wild-type strain. Both ratios were generally constant in each experiment irrespective of the variation in the absolute levels of each parameter, indicating that the chromatin preparations used herein are of consistent quality (Tables 1 and 2).
Table 1.
MNase sensitivity (20 units) in premeiosis (t = 0 h)
| Mutant | Number of data
|
Total DNA cleaved (hot spots), %
|
Ratio, mutant/wt
|
||||
|---|---|---|---|---|---|---|---|
| ARG4 | CYS3-I | CYS3-II | Mutant | wt | Hot spots | Control sites | |
| Type 1 | |||||||
| mre2 | 1 | 1 | 1 | 18.7 | 13.2 | 1.7 ± 0.4 | 1.1 ± 0.3 |
| mre11 | 1 | 2 | 2 | 12.3 | 7.6 | 1.7 ± 0.4 | 1.0 ± 0.1 |
| Type 2 | |||||||
| rad50Δ | 1 | 2 | 2 | 7.6 | 5.5 | 1.3 ± 0.3 | 1.0 ± 0.1 |
| xrs2 | 1 | 2 | 2 | 2.9 | 1.9 | 2.4 ± 1.8* | 1.1 ± 0.1 |
| rad50S | 1 | 1 | 1 | 7.7 | 7.5 | 1.0 ± 0.2 | 1.0 ± 0.0 |
| mre11-rad50Δ | 1 | 2 | 2 | 10.6 | 7.7 | 1.4 ± 0.1 | 1.0 ± 0.1 |
Each mutant was analyzed as described in Fig 2. For all mutants, MNase sensitivity was assayed at both the ARG4 and CYS3 DSB hot spots and at a control non-DSB hypersensitive site (Fig. 1 D and E). The data presented are the averages of MNase digestion levels (i.e., percentage of the total lane intensity including values for an unbroken parental fragment) obtained in several experiments using 20 units of MNase; qualitatively analogous results are obtained by using higher and lower MNase levels (Fig. 2 and data not shown). Absolute MNase sensitivity at hot spots represents an average of results at ARG4 and CYS3. Thus, these values are meaningful in comparison of the mutant and the corresponding wild-type control (wt) in each line. Ratio of the sensitivity in mutants to wild type is the average (±SD) of the ratio taken in each experiment (ratio taken in each experiment and then averaged). The experimental variation in the xrs2 data (marked by the asterisk) probably reflects subtle differences in procedures as performed by different experimenters.
Table 2.
MNase sensitivity (20 units) in meiosis (t = 4 h)
| Mutant | Total DNA cleaved (hot spots), %
|
Ratio, mutant/wt
|
Ratio to premeiosis, hot spots
|
||||
|---|---|---|---|---|---|---|---|
| Mutant | wt | Hot spots | Control sites | Mutant | wt | Mutant* | |
| Type 1 | |||||||
| mre2 | 16.8 | 31.1 | 0.6 ± 0.1 | 1.0 ± 0.3 | 1.0 ± 0.2 | 2.9 ± 0.6 | 1.0 |
| mre11 | 17.7 | 21.5 | 0.8 ± 0.1 | 1.0 ± 0.1 | 1.4 ± 0.3 | 2.9 ± 0.5 | 1.4 |
| Type 2 | |||||||
| rad50Δ | 14.9 | 10.9 | 1.3 ± 0.2 | 0.9 ± 0.1 | 2.2 ± 0.6 | 2.1 ± 0.5 | 3.0 |
| xrs2 | 6.9 | 4.1 | 2.5 ± 1.9** | 1.0 ± 0.1 | 2.3 ± 0.3 | 2.3 ± 0.5 | 2.9 |
| rad50S | 16.2 | 15.3 | 1.1 ± 0.1 | 1.1 ± 0.0 | 2.3 ± 0.6 | 2.2 ± 0.5 | 3.0 |
| mre11-rad50Δ | 12.9 | 17.7 | 0.7 ± 0.2 | 1.0 ± 0.0 | 1.2 ± 0.2 | 2.5 ± 0.9 | 1.4 |
Cells were sampled at t = 4 h. Data analysis and number of experiments are as shown in Table 1 and Figs. 1–3. Ratios of the sensitivity in mutants to wild type (wt) are calculated as in Table 1. Ratios of meiotic to premeiotic sensitivity are calculated by using premeiotic data in Table 1. Premeiotic to meiotic ratios for mutants* were calculated by the following formula: (premeiotic to meiotic ratio for mutants)/(premeiotic to meiotic ratio for wild-type) × 2.9, the premeiotic to meiotic ratio for wild-type in the mre2 experiment). The experimental variation in the xrs2 data (marked by the double asterisk) probably reflects subtle differences in procedures as performed by different experimenters. Data are the average ± SD.
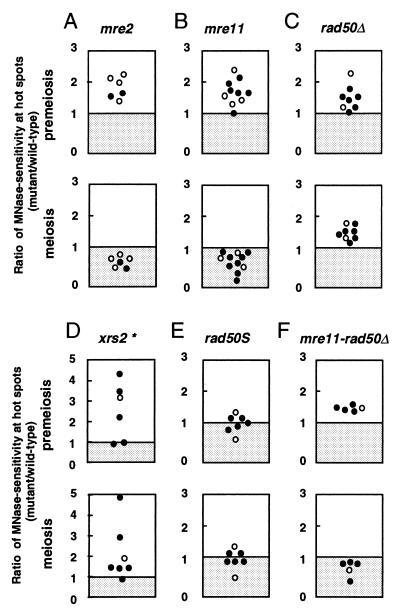
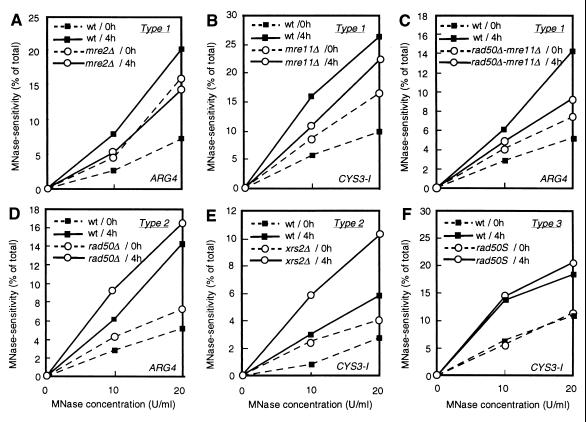
The mutant effects are summarized in Tables 1 (premeiosis) and 2 (meiosis) and illustrated in Figs. 2 and 3. In Fig. 2, each dot represents the ratio of mutant MNase sensitivity to wild-type sensitivity obtained with the wild type defined as unity for each experiment. The data in Tables 1 and 2 represent the average of the ratio of mutant to wild-type MNase sensitivity in each experiment using MNase at 20 units/ml, along with the average of absolute values at ARG4 and CYS3. We found that premeiotic MNase sensitivity at DSB hot spots in all mutants was slightly but significantly higher than in the wild-type control. The ratio of mutant to wild-type MNase sensitivity at DSB hot spots was reproducibly more than 1.0 (Fig. 2 and Table 1). On the other hand, the ratio of mutant to wild-type MNase sensitivity at a site without DSBs (control sites) was always around 1.0 (Table 1), suggesting that the mutant effects are specific to DSB sites. We found no significant difference between ARG4 and CYS3 loci (Fig. 2). We also confirmed that these mutant effects are not dependent on the concentration of MNase used (Fig. 3). Although all four disruptants displayed higher premeiotic MNase sensitivity at DSB sites, it remains to be examined whether the four strains are identical or significantly different under the premeiotic conditions. On the other hand, no apparent premeiotic effect on MNase sensitivity could be detected in the rad50S mutant (a DSB-proficient separation of function RAD50 mutant) as compared with wild type (Figs. 2 and 3 and Table 1).
Figure 2.
Ratio of mutant MNase sensitivity to that of the wild-type control. Each mutant was analyzed in several experiments using various concentrations (7, 10, and 20 units/ml) of MNase. In each experiment, two or three mutant cultures were analyzed in parallel with a wild-type control culture on the same day. Ratio of mutant MNase sensitivity to wild-type sensitivity at ARG4 (○) and CYS3 (•) DSB sites were plotted. Only vertical axes (ratio of mutant to wild type) are meaningful. (A) mre2. (B) mre11. (C) rad50Δ. (D) xrs2. (E) rad50S. (F) mre11-rad50Δ. (Upper) Premeiotic data. (Lower) Meiotic data. The experimental variation in the xrs2 data (marked by the asterisk) probably reflects subtle differences in procedures as performed by different experimenters.
Figure 3.
Quantitative comparison of MNase hypersensitivity. MNase hypersensitivity (expressed as percentage of total lane intensity) at ARG4 (A, C, and D) and CYS3 site I (B, E, and F) in mre2 (A), mre11 (B), mre11 rad50Δ (C), rad50Δ (D), xrs2 (E), and rad50S (F) is plotted as a function of MNase concentration. The mutant and wild-type cultures were always analyzed in parallel on the same day. For an accurate evaluation of the mutant phenotypes, all mutant data are indicated with wild-type data for an internal standard taken on the same day. Solid symbols, wild type; open symbols, mutants. Dashed lines, 0 h (premeiosis); solid lines, 4 h (meiosis). Note that the mutants are classified into groups (types 1, type 2, and rad50S, as indicated) and that the mutant effects are independent of the MNase concentration.
The mre11, rad50, xrs2, and mre2 Mutants Define Two Aberrant Meiotic Chromatin Configurations at DSB Sites.
We have reported (37) that MNase sensitivity at DSB hot spots increases significantly during meiosis in a wild-type strain background. In the present study, we detected a similar increase (2.1- to 2.9-fold) in MNase sensitivity at DSB hot spots during meiosis in wild-type rapidly sporulating cells of the SK1 background (Table 2). Parallel quantitative data analysis of the mutants reveals that the four mutants examined herein differ from wild type and fall into two distinct categories (Figs. 2 and 3 and Table 2).
For mutants of the type 1 class, which consists of mre2 and mre11, the final level of MNase sensitivity in meiotic cells is lower than that observed in wild-type cells at t = 4 h (Table 2) and at t = 6 h (data not shown). The ratios of mutant to wild-type MNase sensitivity are 0.6 ± 0.1 (n = 3) and 0.8 ± 0.1 (n = 5) in the mre2 and mre11 mutants, respectively. In Table 2, the difference in meiotic MNase sensitivity between mre11 and wild-type strains seems small (0.8 ± 0.1). However, we believe that this difference is significant, from the scattered plot (Fig. 2) that also includes more data of the digestion at 7–10 units/ml. Because at a non-DSB site, this ratio remains close to unity (1.0 ± 0.3 and 1.0 ± 0.1 in the mre2 and mre11, respectively), this lack of a meiotic increase in MNase sensitivity in the mutant diploids is likely to be specific to DSB regions. The progression of meiosis I in the type 1 mutants is similar to that in wild type, as revealed by patterns of nuclear staining with 4′,6-diamidino-2-phenylindole (K.O., M.F., and T.S., unpublished observation). Thus, these results are probably not due to meiotic asynchrony or to a delay in meiotic division I in the type 1 mutants.
In the type 2 mutants, rad50Δ (null allele) and xrs2, the final meiotic levels of MNase sensitivity at DSB sites are higher than those in the wild-type strain (Table 2). The ratio of mutant to wild-type MNase sensitivity in meiosis is reproducibly higher than one (Fig. 2), whereas sensitivity at a non-DSB site remains unchanged (the ratio of mutant to wild type is 0.9–1.0). Thus, the higher meiotic sensitivity of the type 2 mutants is also specific to DSB hot spots. These values indicate that the type 2 mutations confer a meiosis-specific increase in MNase sensitivity (2.2- to 2.3-fold) as also observed in wild type.
In contrast, the non-null rad50S allele shows almost wild-type levels of MNase sensitivity at the ARG4 and CYS3 DSB hot spots in both premeiotic and meiotic cells (Tables 1 and 2 and Fig. 2).
mre11 Is Epistatic to rad50.
To examine the relationship between the type 1 and the type 2 mutations, we examined a rad50Δ-mre11∷URA3 double mutant. In premeiosis, the two single mutants and the double mutant exhibited similarly elevated levels of MNase sensitivity (Figs. 2 and 3 and Table 1). On the other hand, in meiosis, the double mutant was phenotypically similar to mre11∷URA3 but not to rad50Δ (Figs. 2 and 3 and Table 2). These results show that for premeiotic chromatin at potential DSB sites, mre11∷URA3 and rad50Δ, which are type 1 and type 2 mutations, respectively, belong to the same epistasis group; in contrast, for DSB sites in meiotic chromatin, mre11∷URA3 (type 1) is epistatic to rad50Δ (type 2).
DISCUSSION
Positioning of MNase-Hypersensitive Sites Is Independent of MRE11, RAD50, XRS2, and MRE2 Functions.
Previous studies performed in the wild-type strain background indicated that meiotic DSB regions are characterized by the preferential accessibility of chromatin, as shown by their DNase I and MNase hypersensitivity in vegetative and meiotic cells (10, 37, 38, 49). In early meiosis, prior to DSB formation, MNase hypersensitivity in DSB regions increases (ref. 37 and this report). The molecular basis of this developmental modification of DNA accessibility in chromatin is unknown. In the present report, we have examined the DNA accessibility in chromatin in four mutants (mre11, rad50, xrs2, and mre2) that are defective in DSB formation. At both the ARG4 and CYS3 loci, we find that these mutations do not affect the formation or the location of MNase-hypersensitive sites. This result suggests that factors essential for the initiation of recombination by DSBs are not necessarily required for the formation of nuclease-hypersensitive sites in chromatin. These sites might be defined primarily by the actions of other proteins such as transcription factors. However, a quantitative analysis of chromatin in mutants defective in DSB formation and repair reveals that the extent of MNase sensitivity differs significantly from that in wild-type strains in both premeiosis and meiosis.
Premeiotic Effects.
We found that mre11, rad50, xrs2, and mre2 disruptions confer a slight increase in MNase sensitivity at DSB hot spots when mutant strains of the rapidly sporulating SK1 background are grown in presporulation medium almost to stationary phase. We refer to SK1 cells under these conditions as “premeiotic,” because under certain nutritional conditions as they approach meiosis all cells rather than a subset are in a state distinct from both the mitotic and meiotic states (53). During premeiosis, it is likely that wild-type cells begin producing at least some meiotic factors. Evidence for the existence of the premeiotic state is provided by analysis of mre2 (20). Mre2 protein has a role in the splicing of the MER2 and other primary transcripts in meiosis. In vegetatively grown wild-type cells, spliced MER2 transcript cannot be detected (17), but a low level of spliced transcript appears during premeiosis (20). It is possible that the splicing of other MRE2-dependent mRNAs also occurs in premeiosis, which may reflect a leaky phenotype of the mre2 mutant in premeiosis.
The enhanced premeiotic MNase hypersensitivity of the four mutants may result from an altered protein composition at DSB hot spots in premeiotic chromatin as discussed later. Alternatively, we consider other less compelling explanations for their effect on chromatin. (i) The enhanced hypersensitivity in the mutants may reflect a difference in the timing of entry into premeiosis and meiosis after exponential growth. This possibility is unlikely, because by 4′,6-diamidino-2-phenylindole staining, we could not detect a significant difference between mutant and wild-type strains in the progression of meiosis up to meiotic division I. (ii) A global reduction in nucleosome density may result in an overall increase in MNase sensitivity. This is also unlikely, because the mutant effects are only detected at DSB hot spots. (iii) It is conceivable that premeiotic effects in mre11, rad50, and xrs2 strains are an indirect consequence of events occurring during vegetative growth (23), because these mutants grow slowly and exhibit a hyperrecombination phenotype (32, 54, 55). This possibility cannot be formally ruled out, although it is difficult to explain why the mutant effects are restricted to DSB MNase-sensitive sites. Future studies of the effects of these mutations on mitotic MNase sensitivity are warranted.
Meiotic Effects.
With respect to their effects on MNase sensitivity at DSBs sites during meiotic prophase, the mutants examined herein fall into two categories. The chromatin of mre11 and mre2 mutants (type 1) is less MNase-sensitive than that of wild-type strains, whereas the chromatin of xrs2 and rad50Δ mutants (type 2) is more sensitive.
The lower MNase sensitivity at DSB hot spots in type 1 mutants suggests that the MRE2 and MRE11 gene products directly or indirectly affect meiotic chromatin at these sites. Although the mre2 and mre11 mutations similarly affect MNase sensitivity, the two cases may not be equivalent. The data in Table 2 show that meiotic chromatin is slightly more sensitive in the mre11 (0.8 ± 0.1) than in the mre2 (0.6 ± 0.1) mutant. Because Mre2 is supposed to have a role in the meiosis-specific splicing of MER2 and other gene transcripts, defects in mre2 might occur more indirectly and at a earlier stage than mre11. This notion needs to be tested by detailed side-by-side comparisons of the meiotic effects in both mutants.
MNase sensitivity at DSB hot spots in type 2 mutants is significantly higher than in wild-type strains. This result indicates that the absence of type 2 gene products confers an aberrant chromatin configuration at DSB hot spots that differs from that found in type 1 mutants. This means that Rad50-Xrs2 and Mre11 can be functionally distinguished. Double mutant analysis reveals that the type 1 mutation mre11∷URA3 is epistatic to a type 2 mutation (rad50Δ), implying that the changes observed in type 2 mutants are dependent upon type 1 functions and that type 1 proteins act earlier than type 2 proteins. Thus, we propose that Mre11 and other type 1 proteins may have more basic structural roles in the assembly of the pre-DSB complex, for example, by binding to DNA and recruiting other factors. However, we cannot rule out a possibility that the mutants in the two classes may have subtle difference also in premeiosis.
Mutant Effects on the Premeiosis/Meiosis Change in MNase Sensitivity at DSB Sites.
The induction of MNase sensitivity at DSB hot spots observed in wild-type cells is only partial in type 1 mutants (Table 2, see Ratio to premeiosis). This defect might reflect the direct or indirect involvement of type 1 proteins in the process(es) required for the premeiosis/meiosis chromatin transition at DSB sites. Alternatively, it could be a consequence of premeiotic effects and two different hypotheses can then be considered. (i) An aberrant premeiotic state (slightly higher sensitivity) might be replaced by a normal meiotic state. Changes in chromatin would then occur substantially during meiosis but not normally. This possibility seems unlikely, because we could not detect any decrease in MNase sensitivity in the mre11 mutant at time points earlier than t = 4 h (data not shown). (ii) The chromatin in type 1 mutants could be trapped in an aberrant premeiotic configuration during meiotic prophase, thereby preventing meiotic changes in chromatin structure. However, such a trapping of chromatin also seems unlikely, because both the mre2 and mre11 mutants can undergo premeiotic DNA synthesis (19). Therefore, we favor the notion that type 1 proteins may be involved in the premeiosis/meiosis chromatin transition.
On the other hand, the induction of MNase sensitivity during meiosis can be detected in type 2 mutants. This suggests that type 2 mutations do not prevent the premeiosis/meiosis transition of chromatin at DSB hot spots. However, the final meiotic level of MNase sensitivity is above the level observed in wild type. We consider several explanations for this observation. (i) The normal meiosis-specific change may be superimposed upon an independent premeiotic effect to produce higher levels of meiotic MNase sensitivity. (ii) A meiosis-specific aberration independent of the premeiotic effect may operate in the mutants. (iii) Premeiotic effects may persist and influence the meiotic levels of MNase sensitivity. Further studies are necessary to discriminate among these hypotheses.
No Significant Effect in rad50S.
The rad50S mutant is phenotypically similar to wild type with respect to premeiotic and meiotic MNase sensitivity. This is consistent with the finding that rad50S is a separation-of-function mutation, in that the Rad50S protein retains many functions. In meiosis, it is able to form DSBs but is defective in their resection (24). It has no strong vegetative phenotype, except for a weak methyl methanesulfonate sensitivity and a tendency to undergo telomere lengthening (36).
What Happens at DSB Sites Before DSB Formation?
The premeiotic and meiotic mutant phenotypes described in the present study can be readily explained if the relevant gene products (possibly Mer2 in the case of the mre2 mutant) act directly rather than indirectly at DSB hot spots that are localized within nuclease-hypersensitive regions. At present, we favor the interpretation that these premeiotic and meiotic effects result from an altered protein composition at DSB hot spots in chromatin. This idea is consistent with previous observations concerning Mre11, Rad50, and Xrs2 (21, 32, 33, 48). Recent data indicate that the Spo11 protein is likely to be responsible for the DNA cleavage (29, 30). How Spo11 gains access to its numerous chromosomal DNA targets within the entire genome (6, 11) is unknown. One possibility is that Spo11 is preferentially directed to a Rad50-Mre11-Xrs2 pre-DSB complex assembled on nucleosome-free regions in chromatin.
Interaction of these proteins at DSB hot spots is compatible with the notion that these regions are sites for interhomolog interactions even in the absence of DSBs (44, 46, 47). The presence of heterology at DSB hot spots causes a decrease in DNase I hypersensitivity in mitosis (49) and also causes a reduction in the frequency of meiotic DSBs (44, 46). These results suggest that interhomolog interactions are established at DSB hot spots at a stage earlier than meiotic DSB formation. Because RAD50 has been implicated in homologous pairing in mitosis (B. Weiner, S. Burgess, and N. Kleckner, personal communication) and premeiosis (48), our results support the involvement of the Rad50/Mre11/Xrs2 proteins in these interactions.
Acknowledgments
We thank K. Kobayashi for assistance with these experiments; and K. Johzuka, V. Rocco, J. E. Haber, and M. Lichten for providing the mre11, rad50, and xrs2 disruptants and rad50S strains, respectively. We are grateful to N. Kleckner for stimulating discussions on the data and sharing data in advance of publication, to D. Carroll for helpful suggestions, and to K. Smith for English corrections. This work was supported by a research grant from the Human Frontier Science Program (RG493/95), a grant for the Biodesign Research Program from The Institute of Physical and Chemical Research, and grants from the Ministry of Education, Science, Culture, and Sports, Japan.
ABBREVIATIONS
- DSB
DNA double-stranded break
- MNase
micrococcal nuclease
References
- 1.Nicolas A, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 2.Sun H, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 3.Cao L, Alani E, Kleckner N. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 4.Schultes N P, Szostak J W. Mol Cell Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White M A, Detloff P, Strand M, Petes T D. Curr Genet. 1992;21:109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- 6.Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G. EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Massy B, Nicolas A. EMBO J. 1993;12:1459–1466. doi: 10.1002/j.1460-2075.1993.tb05789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyon C, Lichten M. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nag D K, Petes T D. Mol Cell Biol. 1993;13:2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu T-C, Lichten M. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 11.Baudat F, Nicolas A. Proc Natl Acad Sci USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichten M, Goldman A S H. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 13.Roeder G S. Proc Natl Acad Sci USA. 1995;92:10450–10456. doi: 10.1073/pnas.92.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinohara A, Ogawa T. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 15.Kleckner N. Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cool M, Malone R E. Mol Cell Biol. 1992;12:1248–1256. doi: 10.1128/mcb.12.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engebrecht J A, Voelkel-Meiman K, Roeder G S. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 18.Rockmill B, Engebrecht J A, Scherthan H, Loidl J, Roeder G S. Genetics. 1995;141:49–59. doi: 10.1093/genetics/141.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ajimura M, Leem S H, Ogawa H. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa T, Ogawa H. Genes Cells. 1997;2:65–79. doi: 10.1046/j.1365-2443.1997.d01-283.x. [DOI] [PubMed] [Google Scholar]
- 21.Johzuka K, Ogawa H. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Game J C, Motimer R K. Mutat Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 23.Farnet C, Padmore R, Cao L, Raymond W, Alani E, Kleckner N. Mechanisms and Consequences of DNA Damage Processing. New York: Liss; 1988. [Google Scholar]
- 24.Alani E, Padmore R, Kleckner N. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 25.Malone R E, Ward T, Lin S, Waring J. Curr Genet. 1990;18:111–116. doi: 10.1007/BF00312598. [DOI] [PubMed] [Google Scholar]
- 26.Esposito R E, Esposito M S. Genetics. 1969;61:79–89. doi: 10.1093/genetics/61.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klapholz S, Waddell C S, Esposito R E. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov E L, Korolev V G, Fabre F. Genetics. 1992;132:651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergerat A, de Massy B, Gadelle D, Varoutas P C, Nicolas A, Forterre P. Nature (London) 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 30.Keeney S, Giroux C N, Kleckner N. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 31.Dolganov G M, Maser R S, Novikov A, Tosto L, Chong S, Bressan D A, Petrini J H J. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond W E, Kleckner N. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nairz K, Klein F. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leach D R, Lloyd R G, Coulson A F. Genetica. 1992;87:95–100. doi: 10.1007/BF00120998. [DOI] [PubMed] [Google Scholar]
- 35.Connelly J C, Leach D R. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 36.Kironmai K M, Muniyappa K. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 37.Ohta K, Shibata T, Nicolas A. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan Q Q, Petes T D. Mol Cell Biol. 1996;16:2037–2043. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Massy B, Rocco V, Nicolas A. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Wu T-c, Lichten M. EMBO J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keeney S, Kleckner N. Proc Natl Acad Sci USA. 1995;92:11274–11278. doi: 10.1073/pnas.92.24.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuno K, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 43.Wu T C, Lichten M. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu L, Kleckner N. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan Q Q, Xu F, White M A, Petes T D. Genetics. 1997;145:661–670. doi: 10.1093/genetics/145.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rocco V, Nicolas A. Genes Cells. 1996;1:645–661. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- 47.Bullard S A, Kim S, Galbraith A M, Malone R E. Proc Natl Acad SciUSA. 1997;93:13054–13059. doi: 10.1073/pnas.93.23.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiner B M, Kleckner N. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 49.Keeney S, Kleckner N. Genes Cells. 1996;1:475–490. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- 50.Lichten M, Goyon C, Schultes N P, Treco D, Szostak J W, Haber J E, Nicolas A. Proc Natl Acad Sci USA. 1990;87:7653–7657. doi: 10.1073/pnas.87.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rocco V, de Massy B, Nicolas A. Proc Natl Acad Sci USA. 1992;89:12068–12072. doi: 10.1073/pnas.89.24.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernardi F, Koller T, Thoma F. Yeast. 1991;7:547–558. doi: 10.1002/yea.320070603. [DOI] [PubMed] [Google Scholar]
- 53.Simchen G, Pinon R, Salts Y. Exp Cell Res. 1972;75:207–218. doi: 10.1016/0014-4827(72)90538-1. [DOI] [PubMed] [Google Scholar]
- 54.Malone R E, Jordan K, Wardman W. Curr Genet. 1985;9:453–461. doi: 10.1007/BF00434050. [DOI] [PubMed] [Google Scholar]
- 55.Ogawa H, Johzuka K, Nakagawa T, Leem S H, Hagihara A H. Adv Biophys. 1995;31:67–76. doi: 10.1016/0065-227x(95)99383-z. [DOI] [PubMed] [Google Scholar]