Abstract
The effective population size (Ne) is an important parameter in ecology, evolutionary biology and conservation biology. It is, however, notoriously difficult to estimate, mainly because of the highly stochastic nature of the processes of inbreeding and genetic drift for which Ne is usually defined and measured, and because of the many factors (such as time and spatial scales, systematic forces) confounding such processes. Many methods have been developed in the past three decades to estimate the current, past and ancient effective population sizes using different information extracted from some genetic markers in a sample of individuals. This paper reviews the methodologies proposed for estimating Ne from genetic data using information on heterozygosity excess, linkage disequilibrium, temporal changes in allele frequency, and pattern and amount of genetic variation within and between populations. For each methodology, I describe mainly the logic and genetic model on which it is based, the data required and information used, the interpretation of the estimate obtained, some results from applications to simulated or empirical datasets and future developments that are needed.
Keywords: effective population size, genetic markers, inbreeding, genetic drift, coalescent
1. Introduction
When the systematic forces of mutation, selection and migration are absent, the genetic properties (such as heterozygosity, number of alleles, allele and genotype frequencies at a locus) of an infinitely large population will remain constant over time. In contrast, such properties of a population with a finite size will change from generation to generation, resulting inherently from the stochastic process of sampling a finite number of gametes during reproduction and survival. The strength of the stochastic process in and thus the extent of the change in genetic properties of a population depend on its effective size, a concept introduced by Wright (1931) and developed by many others, mainly Crow & Kimura (1970). The stochastic changes of the genetic properties are slower in populations with larger effective sizes than those with smaller effective sizes.
The effective population size (Ne) is defined as the size of an idealized Wright–Fisher population (Fisher 1930; Wright 1931), which would give the same value of some specified genetic property as in the population in question (Crow & Kimura 1970). Depending on the genetic property of interest, therefore, different concepts of Ne have been proposed such as the inbreeding effective size, variance effective size, eigenvalue effective size, mutation effective size and coalescent effective size (Ewens 1982; Gregorious 1991; Caballero 1994; Whitlock & Barton 1997; Charlesworth et al. 2003). The most widely used concepts are inbreeding effective size (NeI), which predicts the rate of decrease in heterozygosity or the rate of increase in homozygosity (inbreeding), and variance effective size (NeV), which measures the variance of change in gene frequency resulting from one generation of genetic sampling. In some complex situations where the demography of a population changes over time or an equilibrium state has not been attained, the two effective sizes at any transitory time can be dramatically different (e.g. Chesser et al. 1993; Wang 1997a,b). However, the two effective sizes are the same for equilibrium populations, or are asymptotically the same for non-equilibrium populations when the long-term (harmonic) mean effective sizes are used (Pollak 2002). In this review, I will not distinguish inbreeding and variance effective sizes and will refer to them collectively as effective size (Ne).
There has been tremendous interest in knowing the effective size of a natural or artificial population in many study areas of population genetics, quantitative genetics, evolution and conservation biology. Effective size, interacting with systematic forces such as mutation, selection, migration and recombination, determines the amount and distribution of genetic variation present in a population. From a retrospective point of view, therefore, Ne helps in explaining the observed extent and pattern of genetic variation in a population, in inferring the evolutionary mechanisms involved in shaping the variation in natural populations, and in understanding the evolution of sex and recombination (Barton & Charlesworth 1998). From a prospective point of view, Ne helps to predict the loss and distribution of neutral genetic variation, the fixation probabilities of beneficial or deleterious alleles (Robertson 1961), and the fitness and survival of a small population (Lynch et al. 1995). Therefore, knowledge of Ne facilitates the designs of efficient artificial selection schemes in plant and animal breeding (e.g. Caballero et al. 1991) and the effective management of populations of endangered species (Frankham 1995; Wang 2004).
In spite of our great interest in this important parameter, Ne is notoriously difficult to estimate. As a result, we know very little about the effective sizes of natural populations. A real population may depart from the idealized Wright–Fisher population in many different ways, and as a result, its Ne can be very different from the census size. In addition to the census size, many other demographic and genetic parameters, such as sex ratio, variance of reproductive success among individuals, mating system and mode of inheritance, affect the Ne of a population. The effects of these parameters on Ne have been formulated, and have been reviewed by Caballero (1994) and Wang & Caballero (1999). In principle, these formulations can be used to estimate the Ne of a population if the relevant demographic parameters can be inferred from the population in question. Unfortunately, however, parameters such as the variance of reproductive success are extremely difficult to estimate for natural populations. Pedigree information can also be used to estimate Ne, but again is rarely available from most natural populations. In the unusual case of a well- studied population so that either of these two approaches is applicable, the estimated effective size is the short-term or current value and has thus limited value in explaining the current amount and pattern of genetic variation of the population, which are the result of genetic drift, mutation and other evolutionary forces over a much longer time-scale.
Currently, the most widely used approaches to estimating the effective sizes of natural populations are those based on the genetic properties of the populations as revealed by various genetic markers. The effective size of a population affects both the amount and distribution over time and over space (within and between individuals and subpopulations) of genetic variation, which can be quantified using suitable genetic markers and used to infer the effective population size. Owing to the rapid development in molecular biology in recent years, a variety of markers and DNA sequences can be determined easily and cheaply for a sample of individuals from various species. Under an appropriate genetic model, statistical methods can be developed to extract information from such marker data to estimate effective size alone or together with other interesting parameters, such as migration rate. This paper reviews the methodologies proposed to estimate Ne from genetic data in the last 30 years, with emphasis on those not touched upon or covered only briefly by two previous reviews (Schwartz et al. 1999; Beaumont 2003a). For each methodology, I will describe mainly the logic and genetic model on which it is based, the data required and information used, the interpretation of the estimate obtained, some results from applications to empirical datasets and future developments that are needed. The technical details of the implementation and computation of the methodologies are largely omitted, because they are sometimes quite complicated and have been described in the original papers cited in this review. Because of the large body of literature in this fast growing research area, some relevant papers may inevitably be overlooked by the review. However, I hope the review captures the main classes of Ne estimation methods available, and provides useful information for empiricists in understanding, choosing and applying these methods to their data analysis and for theoreticians in improving current and developing new methods.
2. Methodologies for estimating effective sizes
(a) Current Ne estimated from heterozygote excess
In the absence of selection, mutation and migration, an infinitely large population with discrete generations will attain Hardy–Weinberg equilibrium by one generation of random mating. At the equilibrium, gene frequencies and genotype frequencies are constant from generation to generation, and there is a simple relationship between genotype frequencies and gene frequencies called the Hardy–Weinberg law. For a finite population, however, the Hardy–Weinberg law is violated because genetic drift generates both chance deviations of genotype frequencies from the expected Hardy–Weinberg proportions (HWP) and a systematic bias due to the discreteness of the possible numbers of different genotypes (Kimura & Crow 1963). The bias is towards a heterozygote excess and homozygote deficiency compared with HWP, by an amount of
| (2.1) |
in a Wright–Fisher population with size N (Crow & Kimura 1970). This bias can be regarded as due to the statistical sampling of a finite number of offspring, and applies to a random sample of offspring when N refers to the sample size.
In a finite diploid population with separate sexes, there are additional chance deviations of genotype frequencies from the expected HWP and a systematic heterozygote excess in the offspring, caused by the genetic drift that occurred in the parental generation. This systematic bias can be regarded as being caused by the process of genetic sampling of finite numbers of male and female parents, resulting in a stochastic difference in gene frequency between male and female parents. Robertson (1965) showed that in an idealized population but with separate sexes with Nm male and Nf female parents, the heterozygote excess in the progeny is
| (2.2) |
where is the effective size of the parental population (Caballero 1994). Note that αp is independent of αo in a population with separate sexes. The former is caused by the drift in the parental population and is thus determined by the Ne of the parental population, while the latter is caused by the drift (or sampling) in offspring and is thus determined by the Ne (or sample size) of offspring.
Equations (2.1) and (2.2) suggest an estimator of effective size using the heterozygote excess observed at some marker loci from a single sample. The estimator derived by Pudovkin et al. (1996) for a single biallelic locus is
| (2.3) |
where , Hexp=2p(1−p) is the expected heterozygosity calculated from the gene frequency (p) observed in a sample of N offspring, and Hobs is the observed heterozygosity in the sample. For mutiallelic loci, D is calculated as the average across alleles per locus, and across loci (Pudovkin et al. 1996; Luikart & Cornuet 1999).
Computer simulation studies indicate that this estimator is little biased, but has very low precision (Pudovkin et al. 1996; Luikart & Cornuet 1999). Applying estimator (2.3) to 10 datasets with known small parental population sizes, Luikart & Cornuet (1999) obtained five Ne estimates that are infinity. Until now, there have been few applications of this estimator to the analysis of real datasets. Because the average heterozygosity excess is reciprocally proportional to Ne, the precision of estimator (2.3) decreases with an increasing true effective size. The estimator is useful only for very small random mating populations when many markers are genotyped from a large sample.
The estimation of Ne based on heterozygosity excess can be improved in several aspects. First, more general equations for heterozygosity excess at diploid autosomal loci and haplodiploid loci (or species) owing to sampling/drift in parents (αp) and offspring (αo) were derived for populations with an arbitrary distribution of reproductive success (Wang 1996). These formulae yield a general and simple relationship between Ne and heterozygosity excess when the covariance between the numbers of male and female offspring per parent is zero, which seems to be plausible for most natural populations. A more general estimator should be developed based on these formulae, noting that the heterozygosity excess observed in a sample is partitioned into αo and αp owing to statistical sampling and drift, respectively. Second, the current estimator makes no weighting among the D values calculated from different alleles and loci, resulting in a potential loss of precision. More appropriately, weights should be applied to different alleles and loci depending on their sampling variances determined by allele frequencies and sample sizes. Third, genetic drift generates not only a heterozygosity excess on average, but also chance deviations of genotype frequencies from the expected HWP. The variance of the deviations should have a simple relationship with Ne and thus can be used for estimating Ne as well. Using information on both the mean and variance of the deviations may potentially improve the precision of Ne estimates substantially.
The assumptions of no mutation and no selection in the heterozygosity excess method are valid in general, because only one generation is concerned and markers are ‘neutral’. The assumption of a single isolated population without immigration is violated in some natural populations. When immigration exists, but is ignored, Ne would be underestimated because in addition to drift, immigration also produces heterozygote excess. The strongest assumption made by the heterozygosity excess method seems to be random mating. When mating is not at random, then the heterozygosity excess generated by drift can easily be overwhelmed by that generated by non-random mating. Although the heterozygosity excess has been quantified in a finite population with non-random mating such as partial selfing or full-sib mating (Wang 1996) and can be used to infer Ne, the proportions of selfing or full-sib mating must be known a priori. In reality, such proportions are at best estimated and their sampling errors could further affect the precision and accuracy of Ne estimated from heterozygosity excess.
(b) Short- or long-term Ne estimated from linkage disequilibrium
Linkage disequilibrium (LD) is the non-random association between alleles at different loci in gametes. It can be produced in principle by a number of factors such as migration, direct or indirect (e.g. hitchhiking) selection, and genetic drift in finite populations. For neutral loci unlinked with selected loci in an isolated population with random mating, LD would come exclusively from genetic drift and can be used to estimate Ne (Hill 1981).
The LD for alleles A and B at two loci is defined as the difference between the frequency of gametes (chromosomes) bearing both alleles (pAB) and the product of the allele frequencies (pApB), DAB=pAB−pApB. The correlation between pA and pB is (Hill & Robertson 1968). If the two loci are neutral and the population has been in isolation with a constant effective size of Ne for sufficiently long time, then rAB will be drawn from an equilibrium distribution determined by Ne and the recombination rate between loci, c. At this equilibrium, we have E(rAB)=0 and
| (2.4) |
approximately (Weir & Hill 1980). When a sample of n chromosomes or individuals are used to estimate allele frequencies and LD, an additional part of V(rAB) owing to sampling, 1/n, should also be included. Hill (1974) showed that the contribution from sampling is the same whether n chromosomes are extracted and identified or n diploid individuals with unknown linkage phase are analysed.
For a number of L loci, there are k=L(L−1)/2 pairs of loci. Denote the correlation, recombination fraction and sample size for the ith pair as ri, ci, and ni (i=1, 2,…,k). Hill (1981) derived a multilocus Ne estimator using the formulation for a single locus above and an approximate variance–covariance matrix of ri2among pairs of loci,
| (2.5) |
where , and ri can be estimated by a variety of methods (Weir 1979). The variance of the estimate is
| (2.6) |
An analysis based on equation (2.6) showed that precise estimates of Ne can be obtained only when the sample size n is large relative to the ratio Ne/γ∼4Nec (Hill 1981) and/or k is large.
Hill (1981) applied his method to the analysis of two datasets on Drosophila melanogaster. The first is a sample of 198 flies collected from a wild population in North Carolina (Langley et al. 1977). For each sampled individual, the second and third chromosomes were extracted and analysed for six and five enzyme loci, respectively. The estimate of Ne from equation (2.5) was negative, indicating that the observed LD is less than that expected from sampling alone and thus the best estimate of Ne is infinitely large. The second dataset is from the Maine cage population of Langley et al. (1978). Three and four enzyme loci on the second and third chromosomes, respectively, were analysed for a sample of 635 to 756 flies, depending on the pair of loci. Applying equations (2.5) and (2.6) yields a Ne estimate of 363 with a standard deviation of 170. The Ne estimate of 363 is below the census size of this cage population, 1000. Applying the LD estimator of Ne to several other empirical datasets yielded plausible results (e.g. Bartley et al. 1992; Ardren & Kapuscinski 2003).
Several issues associated with the LD estimator of Ne need further consideration. First, the estimator was derived using a number of approximations. For example, equation (2.4) is not a good approximation when gene frequencies are very close to zero or one (Hill 1981; Hudson 1985). A simulation study is necessary to investigate how reliable these approximations are, whether the estimator is biased or not, and how accurate the estimate of variance as formulated in equation (2.6) is. Second, the estimator assumed an isolated equilibrium population with a constant Ne. The assumption may not be tenable for some natural populations, especially in the long run. Hill (1981) showed that more information comes from pairs of more tightly linked loci, and therefore one may choose closely linked markers to better estimate Ne from LD. However, the tighter the linkage between markers, the longer the time it requires the LD to reach an equilibrium distribution. In other words, the LD for more tightly linked markers observed in the current population would reflect (remember) a longer period of the past demographic history of the population, and thus is more probably affected by factors such as founder event, migration and fluctuation in population size (Hill 1981). On the positive side, one may estimate Ne separately from loosely linked loci and from tightly linked loci to obtain some information on the demographic history of the population (Hill 1981). On the negative side, it is difficult to interpret the exact meaning of the Ne estimate obtained. For example, a smaller estimate of Ne from tightly linked markers than that from loosely linked markers may be a result of a founder event, a bottleneck in population size in the remote past, a gradual growth in population size, an immigration or a hybridization event in the past or any combination of these events. Third, the LD estimator uses information on pairs of loci, and additional information could be exploited from groups of three, four or more loci. Although the LD among three or more loci generated by drift in a finite random mating population was investigated by Hill (1976), the results are less precise than those for pairs of loci and thus are considered of no practical value in estimating Ne (Hill 1981). Fourth, the estimator was derived assuming biallelic loci. At present, highly polymorphic markers that may have scores of alleles per locus, such as microsatellites, are widely available. One possible way to use multiallelic markers in the LD estimator is to consider, in turn, each allele and bin all the other alleles at a locus, as suggested by Waples (1991). This may be plausible for unlinked loci. How this approximate treatment affects the accuracy and precision of the LD estimator needs further investigation.
Recently, Hayes et al. (2003) developed a method to use the LD information from multiple densely spaced markers on a chromosome segment for inferring the Ne at different time points in the past. They proposed a novel multilocus measure of LD, the chromosome segment homozygosity (CSH), which is defined as the probability that two homologous chromosome segments drawn at random from the population are from a common ancestor without intervening recombination. The CSH cannot be observed directly from marker data but can be inferred from marker haplotype (for the segment) frequencies and marker frequencies, both being observable from a sample of individuals. The expectation of CSH in a population with a constant Ne is the same as that of r2 in equation (2.4), which is approximately 1/(4Nec+1) for small values of c, the recombination fraction or the length of the chromosome segment in Morgans (Hayes et al. 2003). When Ne changes linearly over time, then the expectation of CSH is ca. 1/(4Ne,tc+1), where Ne,t is the effective population size at t=1/2c generations ago in the past (Hayes et al. 2003). Therefore, CSHs for chromosome segments of different lengths (c) can be used to estimate the Nes at different generations in the past. Applying this method to a human haplotype dataset including 24 single nucleotide polymorphisms (SNPs) and 2 microsatellites in a 1 cM region, Hayes et al. obtained an estimate of Ne of about 5000 at about 2000 generations ago, using short lengths of haplotypes, and of about 15 000 at about 182 generations ago, using long lengths of haplotypes. The result suggests an exponential growth of the human population in the past, which seems to be plausible. Hayes et al. also applied their method to a dataset comprising 16 microsatellites in a 65 cM segment on chromosome 20, sampled from 264 Australian Holstein–Friesian cows. The estimated Nes are 250 and 1000 using CSH at large and small lengths, respectively. This suggests a decline in Ne for the population, which again seems to be compatible with the known breeding history of this breed.
There is still room to refine the CSH method. For example, the recent Ne using large chromosome segments tends to be overestimated, suggesting that equation (2.4) may not be a good approximation for larger segments in estimating the Ne in the more recent past. Further, sampling effects due to small sample sizes need to be accounted for; otherwise, like LD for a pair of loci, the effect of Ne,t on CSH will probably be swamped by sampling effects. Mutations, which are ignored by Hayes et al. (2003), may not be a problem for estimating the recent Ne using large chromosome segments, but could have a measurable effect on estimating the Ne hundreds or thousands generations into the past using very small segments.
The above LD methods using either pairs of loci or segments of chromosomes assume an isolated population without immigration. The assumption may not be tenable for some natural populations, especially over long time-intervals. Immigration could lead to underestimation of Ne from the LD methods, because the LD generated by migration is falsely regarded as that produced by drift. Vitalis & Couvet (2001) proposed an estimator that can disentangle migration from drift as sources of LD and thus can estimate both simultaneously. Under the infinite island model (Wright 1951), the Fst for a focal population can be estimated from single locus genotype frequencies and gene frequencies observed from a sample. At equilibrium under migration and drift, the expected value of Fst is ca 1/(1+4Nem), where Ne is the effective size and m is the immigration rate for the focal population. To quantify the non-random association of alleles between a pair of loci, Vitalis & Couvet (2001) defined the parameter of ‘within-subpopulation identity disequilibrium’ as the excess of two-locus identity probabilities over the product of single-locus identity probabilities among individuals within subpopulations. This parameter (denoted by ), when standardized in a way similar to rAB in equation (2.4), can be estimated from two-locus genotype frequencies and gene frequencies observed in a sample. The expected value of is a known function of one- and two-locus identity probabilities, which, in turn, are determined by parameters m and Ne (Vitalis & Couvet 2001). From these known relations and the estimated values of Fst and , one can obtain separate estimates of m and Ne. Simulations show that in general the estimator could return reasonably good estimates of m and Ne when 50 individuals are sampled and 8 or more microsatellites are genotyped, if Ne is not large (<100).
The simulations also indicate that Ne is generally underestimated, especially when mutation is assumed to follow the k-allele model (Vitalis & Couvet 2001). The method relies on the infinite island model under drift and migration equilibrium, which requires a constant population size and migration rate over many generations. When Ne is large, then the one- and two-locus identity disequilibria generated by drift would be too small to allow an accurate estimate of Ne. In such a case, use of closely linked markers can increase the power, but a much larger number of generations would be necessary for the equilibrium to be reached.
(c) Short-term Ne estimated from temporal samples
In general, the allele frequency of a population changes over time owing to either the systematic forces of mutation, selection and migration or the stochastic force of genetic drift, or both. When all the systematic forces are excluded, the observed change in allele frequency comes solely from genetic drift and can thus be used to infer how strong the drift is or how large the Ne of the population is. The so-called ‘temporal methods’ for estimating Ne are based on this logic, and were proposed by Krimbas & Tsakas (1971) and subsequently developed by many others (e.g. Nei & Tajima 1981; Pollak 1983; Waples 1989).
The basic protocol of the approach is as follows. Suppose we have an isolated random mating population with discrete generations and we wish to measure its (average) Ne during a certain period of time. Two samples of individuals can be taken at random from the population, the first sample at the beginning (generation 0) and the second sample at the end (generation t) of the period of time. The two samples are then genotyped for a number of neutral markers to estimate allele frequencies, which are used to estimate the standardized variance in the temporal changes of allele frequency, F. The estimated F, , is thus contributed by both sampling and genetic drift. When the sampling effect is accounted for, reflects the strength of genetic drift and, in expectation, is reciprocally proportional to Ne. An estimate of Ne can then be obtained from .
In the above protocol, the main difficulty comes from estimating F and finding its expectation. Several estimators of F were available, among which a widely used one was developed by Nei & Tajima (1981),
| (2.7) |
where xi and yi are the observed frequencies of allele i at a locus with k alleles in the first and second samples, respectively. For multiple loci, is calculated as the average of single locus estimates. The expectation of depends on the sampling scheme that is used to sample genes from the population. Nei & Tajima (1981) distinguished two sampling schemes.
Scheme 1 assumes that the population's census size (when samples are taken), N, is equal to its Ne, that allele frequencies are determined by sampling Sj (j=0 and t for the first and second sample, respectively) individuals out of N, and that sampling at any generation does not affect population allele frequencies and Ne. The latter assumption holds when individuals are sampled after reproduction or when they are returned to the population after examination of genotypes. The exact expectation of under this sampling scheme is difficult to obtain, but an approximation was derived by Nei & Tajima as . Using as its expectation, we can therefore obtain
| (2.8 |
Scheme 2 assumes that N (when samples are taken) is much larger than Ne, and that individuals for determining allele frequencies and those for generating the next generation are sampled separately from the population of N individuals. Under this sampling scheme, approximately (Nei & Tajima 1981). When N≫Ne, the estimator of Ne is
| 2.9) |
The uncertainty of can be assessed from that of . It was shown that follows roughly the χ2 distribution (Lewontin & Krakauer 1973; Nei & Tajima 1981). When n loci are used in the estimation, follows roughly a χ2 distribution with kI degree of freedom, where and ki is the number of alleles at locus i. The values that give the 2.5 and 97.5% cumulative probabilities can be obtained from this χ2 distribution, which are then used to determine the 95% confidence limits of Ne. In practice, the confidence interval determined by this procedure is a slightly overestimate.
Recently, probability methods have been developed to improve the estimates of Ne from temporal samples. Williamson & Slatkin (1999) proposed a likelihood framework to estimate Ne and its change over time, using two or more samples of biallelic markers. The work was extended by Anderson et al. (2000), who described a Monte Carlo approach to computing the likelihood with data on multiallelic markers. Their algorithm, using importance sampling, is highly computationally demanding. Although it produced reasonably good estimates of Ne with small Monte Carlo variance when applied to small problems, it failed to converge when applied to data involving loci with many alleles (Anderson et al. 2000). Wang (2001) proposed a method to calculate the likelihood for multiallelic markers, which is computationally very efficient and applies to any number of alleles per locus. This method transforms a k-allele locus into k biallelic ‘loci’, each having one of the k alleles with all the other alleles pooled. The overall log-likelihood is approximated by the sum of the log-likelihoods across the biallelic ‘loci’ multiplied by the factor of (k−1)/k to account for the dependence of the k ‘loci’. This treatment reduces to the exact likelihood given by Williamson & Slatkin when markers are biallelic (k=2), and yields indistinguishable results for in terms of accuracy and precision from those from the exact likelihood method when markers are tri-allelic. Note that the strength of dependence of allele frequencies at a locus decreases with k, and therefore k=2 and 3 are the worst possible cases for this approximate treatment of multiallelic loci.
The changes in allele frequency in a population can be modelled by a forward probabilistic approach, as adopted by the likelihood methods above, or by a backward coalescent approach (Berthier et al. 2002; Beaumont 2003b; Laval et al. 2003). These coalescent-based methods are Bayesian, usually resorting to Markov chain Monte Carlo (MCMC) to approximate the posterior distribution of Ne. One potential advantage of the coalescent approach is its computational efficiency when sample size is small but Ne is very large.
The probabilistic approaches have several advantages over moment estimators. First, they generally have higher accuracy and precision than moment methods, as verified by several extensive simulation studies (e.g. Wang 2001; Berthier et al. 2002; Tallmon et al. 2004). Moment estimators tend to overestimate Ne when genetic drift is strong (measured by t/2Ne) and when markers with high allelic diversity are used. In such cases, some low-frequency alleles observed in the first sample are absent from the second sample. Moment estimators implicitly assume that these alleles are lost from the population exactly at the tth generation, while in reality they may be lost one or more generations before the tth generation. Because of this, the variance of the change in allele frequency is underestimated and Ne is overestimated. Furthermore, several approximations were made in deriving equations (2.8 and 2.9) or similar moment estimators, resulting in further bias and imprecision, especially when the sampling interval is short (t<3; Nei & Tajima 1981). Second, likelihood methods naturally weigh information optimally. When there are many temporal samples, for example, the relative information content in different sampling intervals will depend on the relative sample sizes and the number of generations between the samples. This difference in information content is automatically taken into account in likelihood or Bayesian methods but is difficult to incorporate into the moment estimators. Third, the underlying demographic model of likelihood methods is flexible and can be easily modified to allow for the estimation of other interesting parameters, such as population growth rate (Williamson & Slatkin 1999; Wang 2001; Beaumont 2003b). The disadvantage of probabilistic approaches is that they require much more computation than moment estimators, and as a result, some of them have not been tested extensively and have difficulty in being extended to more complicated demographic models.
All the above-mentioned temporal methods assume an isolated population without immigration. However, this assumption may not be valid for most populations in the real world, which are connected through gene flow in the forms of gametes and/or individuals. Furthermore, migration, even occurring at an extremely low rate, can substantially alter the genetic makeup of a population and its changes over time. Therefore, current temporal methods can considerably bias estimates of Ne for populations with immigration. Recently, Wang & Whitlock (2003) extend previous moment and maximum-likelihood methods to allow the joint estimation of Ne and migration rate (m) using genetic samples taken from different populations and time. It is shown that, compared with genetic drift acting alone, migration results in changes in allele frequency that are greater in the short term and smaller in the long term, leading to under- and overestimation of Ne, respectively, if it is ignored.
Temporal methods have been developed mainly for populations with discrete generations. However, long-lived species typically have overlapping generations and samples from them generally contain individuals of different age/sex groups. In principle, the basic methods for discrete generations should apply approximately to populations with overlapping generations, when individuals are sampled representatively (proportionately) from all age/sex classes of the population with an interval of one or preferably more generations (Nei & Tajima 1981). Although prolonging the sampling interval is possible, representative sampling is perhaps not realistic in many cases. First, there might be little information about the composition (distributions of individual age/sex) of the population in question before samples are taken. Second, individuals of different ages/sexes may have different behaviour and habitat preferences, and may not even coexist in the same region, making identification of the appropriate biological population and obtaining a representative sample difficult or virtually impossible (Jorde & Ryman 1995). Even if representative sampling and a long sampling interval have been achieved, the methods assuming discrete generations do not use the information fully in samples from an age-structured population, and result in low-precision estimates. This is because an age-structured population does not constitute a homogeneous breeding unit and different age/sex classes are genetically correlated (Hill 1979). Realizing the above problems, Jorde & Ryman (1995) developed a moment estimator of Ne applicable to populations with overlapping generations, based on the ideal model of a fixed age/sex structure and a constant number of individuals of each age/sex class (Felsenstein 1971; Hill 1972, 1979). The main difficulty in applying their moment estimator to populations with overlapping generations is how to group the data into discrete classes appropriately. A sample now contains individuals from several different age/sex classes, so the temporal changes in allele frequencies can be measured in many different ways through grouping the data differently. For example, allele frequencies can be compared between sampling years within the same age class or between age classes within a single sample, or between cohorts (individuals born in the same year) regardless of the ages and samples the individuals come from. Indeed, there are many different ways data can be grouped, and the particular grouping that is the best and how to combine estimates of Ne obtained from different groupings are not immediately apparent. A likelihood method could solve this problem, so that the available data from different age/sex classes in multiple samples can be optimally used to give a single best estimate of Ne.
(d) Long-term Ne estimated from current genetic variation
The amount and pattern of genetic variation in a current population are shaped by the long-term interaction of evolutionary forces of selection, migration, drift, and mutation. In the simple situation of an isolated population with a constant Ne, the genetic variation at a neutral locus is determined by the input from new mutations and the loss from genetic drift. At drift–mutation equilibrium, the amount of genetic variation is constant, determined by a single quantity θ=4Neu for a diploid population, where u is the rate of mutation per generation per DNA sequence or locus. When a sample of individuals is taken from the population and examined for some neutral markers, the genetic variation revealed by these markers can then be used to infer θ. When independent information about u is available, Ne can be estimated from the estimate of θ. When u is unknown, we can still get estimates of the relative Nes of different populations when the same markers and methods are used to estimate their θs.
(i) DNA sequences
DNA sequences represent the highest level of genetic resolution and allow the development of powerful statistical approaches to the inference of population parameters. Different statistics have been proposed to measure the extent of genetic variation at the DNA level (Nei 1987). For a number of n DNA sequences sampled at random from a population, the genetic variation can be measured by the number of segregating nucleotide sites (S) among the sequences, the average number of nucleotide differences between two sequences (Π) and the number of alleles (i.e. different haplotypes; K). An estimate of θ can be obtained from each of the three measurements, assuming a random mating population at drift–mutation equilibrium without selection, immigration and recombination among sites within a sequence. Under these assumptions and the infinite-sites model, which assumes that the number of nucleotide sites on a non-recombining sequence is so large that each new mutation occurs at a site that has not been mutated before (Kimura 1969), Watterson (1975) derived the mean and variance of S,
| (2.10) |
| (2.11) |
where for i=1, 2. Watterson's estimator, θW, can be obtained from equation (2.10) as
| (2.12) |
with sampling variance obtained from equations (2.10) and (2.11) as
| (2.13) |
Under the same assumptions and mutation model, the mean (Watterson 1975) and variance (Tajima 1983) of Π are
| (2.14) |
| (2.15) |
An estimator of θ, known as Tajima's estimator θT, is thus given by Π, with sampling variance given by equation (2.15).
Under the same assumptions but the infinite-allele model, which assumes that each mutation creates a new allele not currently present in the population (Kimura & Crow 1964), Ewens (1972) showed that the mean of K is given by
| (2.16) |
Ewens' estimator of θ, θE, can then be obtained by solving equation (2.16) for a given number of alleles observed in a sample. The sampling variance of θE is (Chakraborty & Schwartz 1990)
| (2.17) |
All three estimators are unbiased under the assumptions and mutation models. However, they use various summary statistics of the sequence polymorphism observed in a sample for estimating θ, making little use of the genealogical relationships among the sampled sequences (Felsenstein 1992). For a sample of n non-recombining sequences from a population evolving according to the neutral Wright–Fisher model, there is a unique (unknown) genealogy specifying the relationships among the sequences. The genealogy consists of n−1 nodes and 2(n−1) branches. The time, ti, during which there are exactly i (i=2, 3,…,n) sequences in the genealogy, is the time (in units of 2Ne generations) required for i sequences to coalesce to i−1 sequences. According to the standard coalescent theory (Watterson 1975; Kingman 1982a,b; Tajima 1983), ti follows the exponential distribution with parameter λi=i(i−1)/2 with mean and variance , and the covariance of ti and tj (i≠j=2, 3,…,n−1) is zero. If the genealogy is known, Ne can be estimated from each ti, and a least-squares method can give the best estimate by combining the n−1 estimates optimally. Unfortunately, the genealogy is unknown but can be inferred from the sequences. In such an inferred genealogy (gene tree), ti and branch lengths are in units of θ or the expected number of mutations, rather than 2Ne, because in the absence of an outside standard, sequences can only give information on the relative lengths of the intervals in terms of the mutational changes that occurred during them. The relationships between ti (or branch lengths) in a gene tree and θ can be exploited to infer θ.
Fu & Li (1993) showed that the minimum variance of the best unbiased estimator of θ is realized when the gene tree of n sequences can be inferred perfectly and is fully used in estimating θ. Under the neutral Wright–Fisher model without recombination, the minimum variance is
| (2.18) |
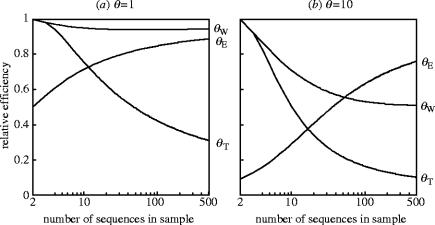
It can be shown that V(θW)=Vmin when θ→0 or n=2; otherwise, V(θW)>Vmin. Similarly, V(θT)=Vmin when n=2; otherwise, V(θT)>Vmin. The relative efficiencies of the three moment estimators indicated by Vmin relative to their respective variances are shown in figure 1. As can be seen, there is much room for improvement on these estimators when θ is not small and the sample size is small (for θE) or large (for θW and θT). θW is generally better than θT, and the difference increases with sample size. The precision of θE changes very fast with sample size, being the worst when n is small and the best when n is large.
Figure 1.
Efficiency of Watterson's estimator θW, Tajima's estimator θT and Ewens' estimator θE relative to the theoretically best estimator with minimum variance Vmin. The θ values are assumed to be 1 (a) and 10 (b).
Fu (1994a,b) developed least-squares estimators of θ using the genealogical relationships provided by the sequence data. For a gene tree of n sequences, let ξi be the total number of mutations on all branches having exactly i (i=1, 2,…,n−1) descendant sequences in the sample. Under the infinite-sites model, it can be shown that and (Fu 1994b). In other words, both estimators are linear functions of ξi with predetermined constant coefficients and therefore are not expected to generate the best estimates in general. Ideally, these ξi should be combined linearly in an estimator using the optical coefficients determined by the least-squares approach, using the variance and covariance structure of ξi. Such an estimator was proposed by Fu (1994a), using the number of mutations on each of the 2(n−1) branches inferred from the gene tree that is reconstructed by the unweighted pair-group method with arithmetic mean. Simulations showed that, under the neutral Wright–Fisher model without recombination, the variance of this estimator can be substantially smaller than those of θW and θT and is always close to Vmin. Using ξi as the primary source of information, Fu (1994b) developed another estimator applicable to populations with recombination or subdivision in the finite island model.
More recent efforts in estimating θ focus on developing probabilistic approaches that take the uncertainty of reconstructed gene trees into account and apply to more complex demographic models and more plausible mutational models. Two distinctive classes of methods are available. One uses the Metropolis–Hastings algorithm to sample a large number of plausible genealogies to calculate the likelihood of the parameter θ given the DNA sequence data (e.g. Kuhner et al. 1995). Under the neutral Wright–Fisher model without recombination and population subdivision, the method of Kuhner et al. (1995) makes maximum-likelihood estimates of θ from DNA sequences evolving under Kimura's (1980) two-parameter model. Their simulation showed that the likelihood estimator is unbiased, and is slightly more precise than Watterson's estimator. Recently, the method has been extended to more complex demographic models, allowing the estimation of θ in the presence of recombination (Kuhner et al. 2000b), population growth or decline in the exponential model (Kuhner et al. 1998) and population subdivision (Beerli & Felsenstein 1999, 2001).
The second class of methods uses coalescent theory to derive recurrent equations for the probability of the polymorphism pattern in a sample, conditional on parameters such as θ. Such recurrent equations are then used either directly or indirectly in a likelihood framework to estimate θ (and other parameters) given data. This class of methods is more general, applying to samples of markers with or without genealogical information. A simple example is about the probability of the number of segregating sites in a sample of DNA sequences. Under the infinite-sites model, the probability of j segregating sites in a sample of n sequences drawn from a population evolving under the neutral Wright–Fisher model is (Tavaré 1984)
| (2.19) |
For a given number of segregating sites observed in a sample of a moderate size, a maximum-likelihood estimate of θ and the confidence intervals can be easily obtained from equation (2.19) (e.g. Wright et al. 2003). In more complicated situations, such recurrent equations can be derived, but are typically unrealistic to solve by direct numerical methods. MCMC approaches are used instead to estimate the probability of the polymorphism pattern in a sample through the recurrent equations, and thus to infer parameters of interest by maximum likelihood (Griffiths & Tavaré 1994a,b, 1995). Recently, the method has been extended to subdivided populations to estimate θ and other parameters such as migration and population growth rates (Bahlo & Griffiths 2000).
The above-mentioned methods assume that the Ne of a population is constant or follows a simple exponential or linear growth model, and that the mutation rate (u) is also constant. Over the long evolutionary scale in which all the sampled sequences coalesce into their most recent common ancestor, it is probable that neither of these assumptions will be met in a real population. To overcome these problems, genetic data should be sampled sequentially from an evolving population, which can then be analysed in a coalescence based likelihood framework to infer Ne and u separately and perhaps their changes over time (Drummond et al. 2002; Seo et al. 2002).
(ii) Microsatellites and other markers
Approaches similar to those for DNA sequence data shown above can be applied to microsatellite and other kinds of markers for estimating θ. The main difference between different markers is the mutational process, not the coalescent process. For microsatellites, the widely used mutation model is the single stepwise mutation model (SSMM; Ohta & Kimura 1973), which assumes that a mutation leads to one repeat unit increase or decrease in allele size with an equal probability. Under this simple mutation model, the variance in allele size expected at the equilibrium between mutation and genetic drift in a Wright–Fisher population without selection and population subdivision is (Moran 1975)
| (2.20) |
and the variance of Vs is (Zhivotovsky & Feldman 1995)
| (2.21) |
Equations (2.20) and (2.21) suggest an estimator of θ using an estimate of the variance in allele size, , from a sample,
| (2.22) |
with sampling variance
| (2.23) |
Estimator θs is unbiased (Xu & Fu 2004), but has a high sampling variance which increases very rapidly with θ, as indicated by equation (2.23).
The genetic variation at a microsatellite locus can also be measured by heterozygosity, H, defined as the probability that two genes drawn at random from a population are of different allelic types. Under the same population and mutation models, the expected heterozygosity at equilibrium is derived (Ohta & Kimura 1973) as
| (2.24) |
If a sample of individuals is taken from the population and examined for a microsatellite, then the population's heterozygosity can be estimated as
| (2.25) |
where k is the number of alleles and pi is the ith allele frequency observed in the sample. A moment estimator of θ can then be obtained from equation (2.24) as
| (2.26) |
θH uses information on both the number and frequencies of alleles, and is generally more precise than θs. Unfortunately, however, θH overestimates θ because of the nonlinear transformation of equation (2.26). Xu & Fu (2004) found that the overestimation is a function of sample size n and θ only, and obtained an empirical regression equation from simulations to correct for the bias. Their simulations showed that θH estimated by equation (2.26) and corrected by their regression equation is unbiased, and has a much smaller sampling variance than θs.
The mutational process for microsatellites can be far more complicated than SSMM assumed above. For example, the rate of mutations leading to expansion may not be equal to that of contraction in allele size (e.g. Chakraborty et al. 1997), a mutation may result in changes of two or more repeat units (Di Rienzo et al. 1994), and the pattern of the mutational processes can differ among loci (Di Rienzo et al. 1998). The simulations of Xu & Fu (2004) showed that both θs and θH estimators are upwardly biased when mutations involving multiple repeat units are allowed to occur, and the upward bias is an increasing function of θ.
Probabilistic methods have also been proposed for estimating θ from microsatellites under SSMM. Nielsen (1997) developed a likelihood method, based on the pioneering work of Griffiths & Tavaré (1994a,b) to estimate θ from microsatellites and to test hypotheses regarding microsatellite evolution. The method was computationally intensive even for a single locus, making accurate estimation using multiple loci difficult. Wilson & Balding (1998) simplified the likelihood computation by treating the (unknown) ancestral allelic states as auxiliary parameters in their MCMC algorithm. They showed by simulations that although a single microsatellite usually does not give enough information for useful inferences of θ and other parameters, several completely linked microsatellites do. Recently, likelihood or Bayesian methods have been proposed to infer θ from microsatellite data under more complex demographic models that allow for population subdivision with migration (Beerli & Felsenstein 1999, 2001) and population growth or decline in the linear or exponential model (Beaumont 1999; Wilson et al. 2003). In parallel, probabilistic methods have been developed for using SNPs in inferring population parameters including θ (Kuhner et al. 2000a; Nielsen 2000; Wilson et al. 2003).
(e) Ancient Ne estimated from current genetic variation
The polymorphisms of markers observed in a sample from the current population can be used to infer the θ or the long-term Ne of the population over the past of the order of Ne generations. For an ancestral species that became extinct in the remote past, it is usually impossible to apply the same approach to estimating its Ne, because genetic polymorphism is generally not observable. However, the genetic polymorphism of an ancestral species (ancestral polymorphism) can be inferred indirectly from that of its two or more descendant species, which can then be used to estimate the ancestral θ or Ne.
A simple method uses orthologous DNA sequences from three closely related species with a known phylogeny (Nei 1987; Wu 1991; Hudson 1992), and is thus called the ‘trichotomy method’. A classical example is the trio consisting of humans, chimpanzees and gorillas, with the first speciation event leading to gorilla lineage and the second speciation event leading to humans and chimpanzees. The principle of the trichotomy method can be illustrated using the above classical example. The genealogy of orthologous DNA sequences sampled from such a trio may or may not be identical to the species phylogeny because of the presence of ancestral polymorphism common to the descendant species. The extent of inconsistency between gene trees and the species tree depends on the amount of ancestral polymorphism and the time-interval (T, in generations) between the two successive speciation events. The larger the Ne of the ancestral species common to humans and chimpanzees relative to T, the greater the proportion, Pdis, of gene trees discordant with the species tree. The two orthologous genes from humans and chimpanzees may either be derived from a common ancestor in the ancestral species during the time-interval T or remain distinct throughout interval T. The probability of the second event is e−t, where t=T/2Ne for autosomal diploid loci, t=T/(Ne/2) for Y-linked or mitochondrial loci assuming a sex ratio of 1, and t=T/(3Ne/2) for X-linked loci. Given the second event, there must exist three distinct ancestral gene lineages before the first speciation event, and three equally probable gene genealogies are possible. One genealogy is consistent with the species tree, and the other two genealogies are discordant with the species tree, resulting in a total discordant proportion of (2/3)e−t. Equating this expected proportion to the observed proportion of discordance among many unlinked autosomal loci, , we therefore obtain
| (2.27) |
The trichotomy method was applied to human and great ape sequence data, yielding estimates for the Ne of the human–chimpanzee ancestral population in the range of 50 000 to 150 000, depending on the datasets used, gene tree reconstruction methods applied and the generation intervals (15 or 20 years) assumed (Ruvolo 1997; Chen & Li 2001).
The trichotomy method is quite simple, but has several drawbacks (Takahata & Satta 2002; Yang 2002). First, the method assumes that gene trees are correctly constructed, so that the inconstancy between gene and species trees comes solely from ancestral polymorphism. In reality, however, gene trees are estimated from sequence data, and thus suffer from sampling errors owing to limited information in the sequence data. The sampling error in reconstructed gene trees is usually quite high, because closely related species are considered and sequences from them are highly similar. The sampling errors in gene tree reconstruction would result in the overestimation of , and thus overestimation of Ne. Second, the trichotomy method is inefficient because only a fraction of the information available from the sequence data is used, while other information (such as branch lengths in gene trees) that is useful for Ne inference is ignored. Third, the interval between the two speciation events, T, was assumed to be known, but in reality it may be unknown or at best is estimated with large sampling errors.
It is obviously desirable to estimate Ne using information on both the topologies and branch lengths of gene trees after accounting for their uncertainties in reconstruction. Takahata (1986) suggested such a method for estimating the Ne of the common ancestors of two closely related species, using a pair of orthologous genes with one from each of the two species. The coalescence time of the two orthologous genes consists of two parts, the species divergence time y and the time x that the two genes coexist in the ancestral species before they coalesce. For any pair of orthologous genes, y is unknown but fixed, while x is variable among pairs of orthologous genes. Both the mean and variance of x among pairs of orthologous genes depend on the Ne of the ancestral species. Using many pairs of orthologous genes, therefore, it is possible to extract information on x and y and, thus, to estimate the Ne of the ancestral species. Such a moment estimator was developed by Takahata (1986), and was later on extended to a full likelihood method and to the case of three extant species, where the Nes of the two extinct ancestral species as well as the two speciation dates were estimated jointly (Takahata et al. 1995; Takahata & Satta 2002). Yang (1997, 2002) extended the method to account for variation in mutation rate among loci, and used the finite-sites model of Jukes & Cantor (1969) to correct for multiple substitutions at the same site. Applying his likelihood and Bayesian methods to the data of Chen & Li (2001), Yang (2002) obtained an estimate of the Ne of the human–chimpanzee ancestral population. The estimate is in the range of 12 000 to 21 000, much smaller than those of previous studies. Rannala & Yang (2003) further extended Yang's Bayesian method, allowing the use of multiple sequences from a species and an arbitrary number of species with a known topology of the species tree.
Another class of methods are suitable for an ancestral species that recently split and diverged into two closely related species. Wakeley & Hey (1997) proposed a moment estimator of the effective size of an ancestral species assumed to have recently been split into two isolated sister species evolving independently. The segregating sites in a sample of orthologous DNA sequences from the two sister species can be partitioned into four mutually exclusive categories. Category one comprises sites that are polymorphic in species 1 but monomorphic in species 2. Category 2 comprises sites that are polymorphic in species 2 but monomorphic in species 1. Category 3 comprises sites that are polymorphic in both species. Category 4 comprises sites showing fixed differences between the two species. Under the neutral infinite-sites model, and assuming a molecular clock and constant effective sizes of the ancestral (NA) and the two descendent species (N1, N2), Wakeley & Hey (1997) derived the expected numbers of sites in the four categories. The four expected numbers turn out to be functions of the four parameters, θi=4Niu (for i=1, 2, A) and τ=2ut, where u is the mutation rate per sequence per generation and t is the divergence time in generations. The four parameters can be obtained by equating the expected to the observed numbers of sites in the four categories and solving the four equations. Simulations show that all the four parameters can be estimated reasonably well with little bias when data on many unlinked loci are available. In the case of few non-recombinant loci, however, the four expected numbers are highly negatively correlated, resulting in a failure of the estimation procedure. Recently, Nielsen & Wakeley (2001) extended the above isolation model to allow asymmetrical migration between the two descendant species and proposed full likelihood and Bayesian methods to estimate all of the six parameters (the above four parameters and two migration rates) jointly. Their methods apply to a single locus without recombination. More recently, the methods were generalized to allow the use of multiple unlinked loci with the same or different modes of inheritance (Hey & Nielsen 2004).
Compared with the long-term Ne of an extant population (species), the Ne of an ancestral extinct species is more difficult to estimate, because the genetic variation observed in current samples has less information about the more remote past. Furthermore, methods for estimating ancestral Ne require more assumptions, and some of them are more likely to be violated than those for estimating the long-term Ne of an extant species. For example, the molecular clock hypothesis typically assumed in methods for estimating ancestral Ne may be a good approximation when the species involved are tightly related and the total divergence time is not very long. Otherwise, different mutation (or substitution) rates must be assumed on different lineages. Because of the long evolutionary history involved, mutation rate heterogeneity both among nucleotide sites within a locus and among different loci might play an important role in interpreting the data. Failure to account for the heterogeneity could lead to an overestimation of x and thus to an overestimation of the ancestral Ne (Takahata & Satta 2002). Currently, mutation rate heterogeneity was taken partially into account by some methods (e.g. Yang 1997, 2002) but was ignored by most others. All of the above methods assume no intragenic recombination during the long period in which the sampled sequences coalesce into their common ancestor. Recombination reduces the variance in coalescence times (x) across loci, resulting in an underestimation of the ancestral Ne when ignored (Takahata & Satta 2002). Wall (2003) proposed a likelihood method based on summary statistics of data, in which he incorporated intragenic recombination but assumed the infinite-sites mutation model with a constant mutation rate among nucleotide sites within a locus and among loci. The method uses a single DNA sequence from each of three or more species, and is computationally very intensive.
3. Discussion
Over the past three decades, we have seen a proliferation of methods developed to estimate the current, past and ancient effective population sizes using genetic marker data. Coupled with the rapid development in molecular biology and computational techniques and facilities, these methods have been widely applied to understanding the demographic history of many species, including humans, and to inferring the evolutionary mechanisms in shaping the genetic variation observed in the current populations. Given the numerous methods available, in designing an experiment and choosing an appropriate method for data analysis, it is fundamental to understand the logic used, the assumptions made, the data and information required and the interpretation of the estimate obtained by each method. This paper provides such an overview of these methods.
It should be noted that different methods have different time-scales on which Ne is measured. The heterozygosity excess methods estimate the effective size of the parental population, the LD methods infer the short- to intermediate-term (mean) effective population size, the length of the term being dependent on the linkage between markers. The temporal methods estimate the harmonic mean effective population size during the period when samples are taken, while the methods considering the mutational process explicitly estimate the long-term Ne in the past on a time-scale of the order of Ne generations. It is important to understand the time-scales behind each method, because natural populations rarely have constant Ne; rather, they are dynamic entities changing sizes and distributions dramatically over time. Therefore, using the same data, different methods could yield considerably different estimates of Ne.
A related issue is the spatial scale over which Ne is measured. In practice, the exact definition of a natural population may be difficult, because it depends on the time-scale concerned, and the migration rate and migration distance of the population. Frogs in a pond, for example, may be regarded as a single random mating population over a short period during which the total immigration rate is sufficiently small, but as a subpopulation connected with other subpopulations over a long period during which the total immigration rate is no longer negligible. The pattern and amount of genetic variation observed in a sample of individuals from the pond can then be used to estimate the short-term (or current) Ne of the pond, and the long-term Ne of the entire metapopulation using appropriate methods (Wang & Whitlock 2003). The time-scale and spatial scale are usually correlated. A small population in an apparently small spatial scale (e.g. a population on an island) may reflect a larger population on a larger spatial scale (e.g. the large continental population from which the island population originates and receives immigrants) in the longer term.
We should bear in mind that any estimation method has a number of assumptions under which the method is derived. In the real world, these assumptions may not be tenable, and violation of one or more of these assumptions may lead to estimates of Ne that make little sense. Random mating, for example, is a critical assumption for the heterozygosity excess methods. When violated, Ne can be biased dramatically. The robustness of each method to the violation of its underlying assumptions has not been evaluated fully, and future work is needed in this respect. Furthermore, the performance and statistical properties of some methods are poorly known, especially those likelihood or Bayesian methods that require intensive computation. Comparison of the methods in precision, accuracy and computational efficiency would facilitate the choice in practical applications.
It is notable that the information available in data is generally not fully used by any single method in estimating Ne. For estimating short-term Ne from temporal samples, for example, the changes in allele frequencies over time is extracted from data and used as information about Ne, while the information about the deviation from HWP and LD in each sample is ignored. Although one can simply use the three pieces of information independently to obtain separate Ne estimates, it is not obvious how to combine them optimally to give a single best estimate. How to use such different pieces of information simultaneously in a single estimator deserves further investigation.
Ne is a parameter summarizing the effects of many other demographic parameters in determining a given genetic property of the population (Caballero 1994). In some cases, such a highly summary parameter is desirable, simplifying both the explanation of the pattern and amount of genetic variation observed in a population and the prediction of the genetic properties (such as loss of variation, fixation probability, changes in fitness of the population) in the future. In other cases, however, it is more helpful to know the details that determine Ne. This is especially true in conservation biology where appropriate management can be exercised only when detailed knowledge of the population is available. For example, a population estimated to have a small Ne can be a result of various causes such as a bottleneck in census size, a biased sex ratio or a large variance in male and/or female reproductive output. Different causes imply different managements suitable to increase the Ne in the future. Unfortunately, current methods estimate a single Ne or, at best, its temporal changes only. To obtain insight into the demographic details of a population, more information is necessary.
Acknowledgments
I wish to thank Bill Hill on his 65th birthday for the memorable years I spent in Edinburgh under his supervision. I thank Mark Beaumont, Brian Charlesworth, Bill Jordan, Peter Visscher, Ziheng Yang and two anonymous referees for helpful comments on the manuscript.
Footnotes
One contribution of 16 to a Theme Issue ‘Population genetics, quantitative genetics and animal improvement: papers in honour of William (Bill) Hill’.
References
- Anderson E.C, Williamson E.G, Thompson E.A. Monte Carlo evaluation of the likelihood for Ne from temporally spaced samples. Genetics. 2000;156:2109–2118. doi: 10.1093/genetics/156.4.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardren W.R, Kapuscinski A.R. Demographic and genetic estimates of effective population size (Ne) reveals genetic compensation in steelhead trout. Mol. Ecol. 2003;12:35–49. doi: 10.1046/j.1365-294x.2003.01705.x. [DOI] [PubMed] [Google Scholar]
- Bahlo M, Griffiths R.C. Inference from gene trees in a subdivided population. Theor. Popul. Biol. 2000;57:79–95. doi: 10.1006/tpbi.1999.1447. [DOI] [PubMed] [Google Scholar]
- Bartley D, Bagley M, Gall G, Bentley M. Use of linkage disequilibrium data to estimate effective size of hatchery and natural fish populations. Conserv. Biol. 1992;6:365–375. [Google Scholar]
- Barton N.H, Charlesworth B. Why sex and recombination? Science. 1998;281:1986–1990. [PubMed] [Google Scholar]
- Beaumont M.A. Detecting population expansion and decline using microsatellites. Genetics. 1999;153:2013–2029. doi: 10.1093/genetics/153.4.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont M.A. Conservation genetics. In: Balding D.J, Bishop M, Cannings C, editors. Handbook of statistical genetics. 2nd edn. Wiley; London: 2003a. pp. 751–766. [Google Scholar]
- Beaumont M.A. Estimation of population growth or decline in genetically monitored populations. Genetics. 2003b;164:1139–1160. doi: 10.1093/genetics/164.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl Acad. Sci. USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier P, Beaumont M.A, Cornuet J.M, Luikart G. Likelihood-based estimation of the effective population size using temporal changes in allele frequencies: a genealogical approach. Genetics. 2002;160:741–751. doi: 10.1093/genetics/160.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A. Developments in the prediction of effective population size. Heredity. 1994;73:657–679. doi: 10.1038/hdy.1994.174. [DOI] [PubMed] [Google Scholar]
- Caballero A, Keightley P.D, Hill W.G. Strategies for increasing fixation probabilities of recessive mutations. Genet. Res. 1991;58:129–138. [Google Scholar]
- Chakraborty R, Schwartz R.J. Selective neutrality of surname distribution in an immigrant Indian community of Houston, Texas. Am. J. Hum. Biol. 1990;2:1–15. doi: 10.1002/ajhb.1310020102. [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Kimmel M, Stivers D.N, Davison L.J, Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl Acad. Sci. USA. 1997;94:1041–1046. doi: 10.1073/pnas.94.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D, Barton N.H. The effects of genetic and geographic structure on neutral variation. Annu. Rev. Ecol. Syst. 2003;34:99–125. [Google Scholar]
- Chen F.C, Li W.H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesser R.K, Rhodes O.E, Sugg D.W, Schnabel A. Effective sizes for subdivided populations. Genetics. 1993;135:1221–1232. doi: 10.1093/genetics/135.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J.F, Kimura M. Harper and Row; New York: 1970. An introduction to population genetics theory. [Google Scholar]
- Di Rienzo A, Peterson A.C, Garza J.C, Valdes A.M, Slatkin M, Freimer N.B. Mutational processes of simple sequence repeat loci in human populations. Proc. Natl Acad. Sci. USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A, Donnelly P, Toomajian C, Sisk B, Hill A, Petzl-Erler M.L, Haines G.K, Barch D.H. Heterogeneity of microsatellite mutations within and between loci, and implications for human demographic histories. Genetics. 1998;148:1269–1284. doi: 10.1093/genetics/148.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J, Nicholls G.K, Rodrigo A.G, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W.J. The sampling theory for selectively neutral alleles. Theor. Popul. Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Ewens W.J. On the concept of effective population size. Theor. Popul. Biol. 1982;21:373–378. [Google Scholar]
- Felsenstein J. Inbreeding and variance effective numbers in populations with overlapping generations. Genetics. 1971;68:581–597. doi: 10.1093/genetics/68.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Estimating effective population size from samples of sequences—inefficiency of pairwise and segregating sites as compared to phylogenetic estimates. Genet. Res. 1992;59:139–147. doi: 10.1017/s0016672300030354. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. Oxford University Press; 1930. The genetical theory of natural selection. [Google Scholar]
- Frankham R. Conservation genetics. Annu. Rev. Genet. 1995;29:305–327. doi: 10.1146/annurev.ge.29.120195.001513. [DOI] [PubMed] [Google Scholar]
- Fu Y.X. A phylogenetic estimator of effective population size or mutation rate. Genetics. 1994a;136:685–692. doi: 10.1093/genetics/136.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X. Estimating effective population size or mutation rate using the frequencies of mutations of various classes in a sample of DNA sequences. Genetics. 1994b;138:1375–1386. doi: 10.1093/genetics/138.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X, Li W.H. Maximum likelihood estimation of population parameters. Genetics. 1993;134:1261–1270. doi: 10.1093/genetics/134.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorious H.-R. On the concept of effective number. Theor. Popul. Biol. 1991;40:269–283. doi: 10.1016/0040-5809(91)90056-l. [DOI] [PubMed] [Google Scholar]
- Griffiths R.C, Tavaré S. Simulating probability distributions in the coalescent. Theor. Popul. Biol. 1994a;46:131–159. [Google Scholar]
- Griffiths R.C, Tavaré S. Sampling theory for neutral alleles in a varying environment. Phil. Trans. R. Soc. B. 1994b;344:403–410. doi: 10.1098/rstb.1994.0079. [DOI] [PubMed] [Google Scholar]
- Griffiths R.C, Tavaré S. Unrooted genealogical tree probabilities in the infinitely-many-sites model. Math. Biosci. 1995;127:77–98. doi: 10.1016/0025-5564(94)00044-z. [DOI] [PubMed] [Google Scholar]
- Hayes B.J, Visscher P.M, McPartlan H.C, Goddard M.E. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res. 2003;13:635–643. doi: 10.1101/gr.387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G. Effective size of populations with overlapping generations. Theor. Popul. Biol. 1972;3:278–289. doi: 10.1016/0040-5809(72)90004-4. [DOI] [PubMed] [Google Scholar]
- Hill W.G. Estimation of linkage disequilibrium in randomly mating populations. Heredity. 1974;33:229–239. doi: 10.1038/hdy.1974.89. [DOI] [PubMed] [Google Scholar]
- Hill W.G. Non-random association of neutral linked genes in finite populations. In: Karlin S, Nevo E, editors. Population genetics and ecology. Academic Press; New York: 1976. pp. 339–376. [Google Scholar]
- Hill W.G. A note on effective population size with overlapping generations. Genetics. 1979;92:317–322. doi: 10.1093/genetics/92.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W.G. Estimation of effective population size from data on linkage disequilibrium. Genet. Res. 1981;38:209–216. [Google Scholar]
- Hill W.G, Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hudson R.R. The sampling distribution of linkage disequilibrium under the infinite allele model without selection. Genetics. 1985;109:611–631. doi: 10.1093/genetics/109.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R.R. Gene trees, species trees and the segregation of ancestral alleles. Genetics. 1992;131:509–512. doi: 10.1093/genetics/131.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde P.E, Ryman N. Temporal allele frequency change and estimation of effective size in populations with overlapping generations. Genetics. 1995;139:1077–1090. doi: 10.1093/genetics/139.2.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T.H, Cantor C.R. Evolution of protein molecules. In: Munro H.N, editor. Mammalian protein metabolism. Academic Press; New York: 1969. pp. 21–123. [Google Scholar]
- Kimura M. The number of heterozygous nucleotide sites maintained in a finite population due to steady flux of mutations. Genetics. 1969;61:893–903. doi: 10.1093/genetics/61.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kimura M, Crow J.F. The measurement of effective population number. Evolution. 1963;17:279–288. [Google Scholar]
- Kimura M, Crow J.F. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J.F.C. The coalescent. Stoch. Proc. Appl. 1982a;13:235–248. [Google Scholar]
- Kingman J.F.C. On the genealogy of large populations. J. Appl. Prob. 1982b;19:27–43. [Google Scholar]
- Krimbas C.B, Tsakas S. The genetics of Dacus oleae. V. Changes of esterase polymorphism in a natural population following insecticide control: selection or drift? Evolution. 1971;25:454–460. doi: 10.1111/j.1558-5646.1971.tb01904.x. [DOI] [PubMed] [Google Scholar]
- Kuhner M.K, Yamato J, Felsenstein J. Estimating effective population size and mutation rate from sequence data using Metropolis–Hastings sampling. Genetics. 1995;140:1421–1430. doi: 10.1093/genetics/140.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner M.K, Yamato J, Felsenstein J. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner M.K, Beerli P, Yamato J, Felsenstein J. Usefulness of single nucleotide polymorphism data for estimating population parameters. Genetics. 2000a;156:439–447. doi: 10.1093/genetics/156.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner M.K, Yamato J, Felsenstein J. Maximum likelihood estimation of recombination rates from population data. Genetics. 2000b;156:1393–1401. doi: 10.1093/genetics/156.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C.H, Ito K, Voelker R.A. Linkage disequilibrium in natural populations of Drosophila melanogaster. Genetics. 1977;86:447–454. [PMC free article] [PubMed] [Google Scholar]
- Langley C.H, Smith D.B, Johnson F.M. Analysis of linkage disequilibrium between allozyme loci in natural populations of Drosophila melanogaster. Genet. Res. 1978;32:215–229. doi: 10.1017/s0016672300018711. [DOI] [PubMed] [Google Scholar]
- Laval G, SanCristobal M, Chevalet C. Maximum-likehhood and Markov chain Monte Carlo approaches to estimate inbreeding and effective size from allele frequency changes. Genetics. 2003;164:1189–1204. doi: 10.1093/genetics/164.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R.C, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, Cornuet J.M. Estimating the effective number of breeders from heterozygote excess in progeny. Genetics. 1999;151:1211–1216. doi: 10.1093/genetics/151.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery J, Burger R. Mutation accumulation and the extinction of small populations. Am. Nat. 1995;146:489–518. [Google Scholar]
- Moran P.A.P. Wandering distributions and the electrophoretic profile. Theor. Popul. Biol. 1975;8:318–330. doi: 10.1016/0040-5809(75)90049-0. [DOI] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York: 1987. Molecular evolutionary genetics. [Google Scholar]
- Nei M, Tajima F. Genetic drift and estimation of effective population-size. Genetics. 1981;98:625–640. doi: 10.1093/genetics/98.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. A likelihood approach to populations samples of microsatellite alleles. Genetics. 1997;146:711–716. doi: 10.1093/genetics/146.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics. 2000;154:931–942. doi: 10.1093/genetics/154.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Kimura M. A model of mutation appropriate to estimate the number of electrophoretic detectable alleles in a finite population. Genet. Res. 1973;22:201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- Pollak E. A new method for estimating the effective population size from allele frequency changes. Genetics. 1983;104:531–548. doi: 10.1093/genetics/104.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E. Eigenvalue effective population numbers for populations that vary cyclically in size. Math. Biosci. 2002;177:11–24. doi: 10.1016/s0025-5564(01)00109-2. [DOI] [PubMed] [Google Scholar]
- Pudovkin A.I, Zaykin D.V, Hedgecock D. On the potential for estimating the effective number of breeders from heterozygote-excess in progeny. Genetics. 1996;144:383–387. doi: 10.1093/genetics/144.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z.H. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A. Inbreeding in artificial selection programmes. Genet. Res. 1961;2:189–194. doi: 10.1017/S0016672308009452. [DOI] [PubMed] [Google Scholar]
- Robertson A. The interpretation of genotypic ratios in domestic animal populations. Anim. Prod. 1965;7:319–324. [Google Scholar]
- Ruvolo M. Molecular phylogeny of the hominoids: Inferences from multiple independent DNA sequence data sets. Mol. Biol. Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- Schwartz M.K, Tallman D.A, Luikart G. Review of DNA-based census and effective population size estimators. Anim. Conserv. 1999;1:293–299. [Google Scholar]
- Seo T.K, Thorne J.L, Hasegawa M, Kishino H. Estimation of effective population size of HIV-1 within a host: a pseudomaximum-likelihood approach. Genetics. 2002;160:1283–1293. doi: 10.1093/genetics/160.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. An attempt to estimate the effective size of the ancestral species common to 2 extant species from which homologous genes are sequenced. Genet. Res. 1986;48:187–190. doi: 10.1017/s001667230002499x. [DOI] [PubMed] [Google Scholar]
- Takahata N, Satta Y. Pre-speciation coalescence and the effective size of ancestral populations. In: Slatkin M, Veuille M, editors. Developments in theoretical population genetics. Oxford University Press; 2002. pp. 52–71. [Google Scholar]
- Takahata N, Satta Y, Klein J. Divergence time and population-size in the lineage leading to modern humans. Theor. Popul. Biol. 1995;48:198–221. doi: 10.1006/tpbi.1995.1026. [DOI] [PubMed] [Google Scholar]
- Tallmon D.A, Luikart G, Beaumont M.A. Comparative evaluation of a new effective population size estimator based on approximate Bayesian computation. Genetics. 2004;167:977–988. doi: 10.1534/genetics.103.026146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. Line-of-descent and genealogical processes, and their applications in population genetics models. Theor. Popul. Biol. 1984;26:119–164. doi: 10.1016/0040-5809(84)90027-3. [DOI] [PubMed] [Google Scholar]
- Vitalis R, Couvet D. Estimation of effective population size and migration rate from one- and two-locus identity measures. Genetics. 2001;157:911–925. doi: 10.1093/genetics/157.2.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J, Hey J. Estimating ancestral population parameters. Genetics. 1997;145:847–855. doi: 10.1093/genetics/145.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J.D. Estimating ancestral population sizes and divergence times. Genetics. 2003;163:395–404. doi: 10.1093/genetics/163.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.L. Deviation from Hardy–Weinberg proportions in finite populations. Genet. Res. 1996;68:249–257. [Google Scholar]
- Wang J.L. Effective size and F-statistics of subdivided populations. 1. Monoecious species. Genetics. 1997a;146:1453–1463. doi: 10.1093/genetics/146.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.L. Effective size and F-statistics of subdivided populations. 2. Dioecious species. Genetics. 1997b;146:1465–1474. doi: 10.1093/genetics/146.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.L. A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet. Res. 2001;78:243–257. doi: 10.1017/s0016672301005286. [DOI] [PubMed] [Google Scholar]
- Wang J.L. Application of the one-migrant-per-generation rule to conservation and management. Conserv. Biol. 2004;18:332–343. [Google Scholar]
- Wang J.L, Caballero A. Developments in predicting the effective size of subdivided populations. Heredity. 1999;82:212–226. [Google Scholar]
- Wang J.L, Whitlock M.C. Estimating effective population size and migration rates from genetic samples over space and time. Genetics. 2003;163:429–446. doi: 10.1093/genetics/163.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R.S. A generalised approach for estimating effective population size from temporal changes in allele frequency. Genetics. 1989;121:379–391. doi: 10.1093/genetics/121.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R.S. Genetic methods for estimating the effective size of cetacean populations. Rep. Int. Whaling Comm. (Special Issue) 1991;13:279–300. [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weir B.S. Inference about linkage disequilibrium. Biometrics. 1979;35:235–254. [PubMed] [Google Scholar]
- Weir B.S, Hill W.G. Effect of mating structure on variation in linkage disequilibrium. Genetics. 1980;95:477–488. doi: 10.1093/genetics/95.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M.C, Barton N.H. The effective size of a subdivided population. Genetics. 1997;146:427–441. doi: 10.1093/genetics/146.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson E.G, Slatkin M. Using maximum likelihood to estimate population size from temporal changes in allele frequencies. Genetics. 1999;152:755–761. doi: 10.1093/genetics/152.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.J, Balding D.J. Genealogical inference from microsatellite data. Genetics. 1998;150:499–510. doi: 10.1093/genetics/150.1.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.J, Weale M.E, Balding D.J. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Stat. Soc. Ser. A. 2003;166:155–188. [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann. Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wright S.I, Lauga B, Charlesworth D. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 2003;12:1247–1263. doi: 10.1046/j.1365-294x.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- Wu C.I. Inferences of species phylogeny in relation to segregation of ancient polymorphisms. Genetics. 1991;127:429–435. doi: 10.1093/genetics/127.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Fu Y.X. Estimating effective population size or mutation rate with microsatellites. Genetics. 2004;166:555–563. doi: 10.1534/genetics.166.1.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.H. On the estimation of ancestral population sizes of modern humans. Genet. Res. 1997;69:111–116. doi: 10.1017/s001667239700270x. [DOI] [PubMed] [Google Scholar]
- Yang Z.H. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics. 2002;162:1811–1823. doi: 10.1093/genetics/162.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky L.A, Feldman M.W. Microsatellite variability and genetic distances. Proc. Natl Acad. Sci. USA. 1995;92:11 549–11 552. doi: 10.1073/pnas.92.25.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]