Abstract
The Drosophila Myb–Muv B (MMB)/dREAM complex regulates gene expression and DNA replication site-specifically, but its activities in vivo have not been thoroughly explored. In ovarian amplification-stage follicle cell nuclei, the largest subunit, Mip130, is a negative regulator of replication, whereas another subunit, Myb, is a positive regulator. Here, we identified a mutation in mip40 and generated a mutation in mip120, two additional MMB subunits. Both mutants were viable, but mip120 mutants had many complex phenotypes including shortened longevity and severe eye defects. mip40 mutant females had severely reduced fertility, whereas mip120 mutant females were sterile, substantiating ovarian regulatory role(s) for MMB. Myb accumulation and binding to polytene chromosomes was dependent on the core factors of the MMB complex. In contrast to the documented mip130 mutant phenotypes, both mip40 and mip120 mutant males were sterile. We purified Mip40-containing complexes from testis nuclear extracts and identified tMAC, a new testis-specific meiotic arrest complex that contained Mip40, Caf1/p55, the Mip130 family member, Always early (Aly), and a Mip120 family member, Tombola (Tomb). Together, these data demonstrate that MMB serves diverse roles in different developmental pathways, and members of MMB can be found in alternative, noninteracting complexes in different cell types.
Keywords: Drosophila, Myb–Muv B, replication, spermatid differentiation, chorion amplification
Coordinating developmentally regulated transcription and replication patterns in metazoans is critical for differentiation of tissue-specific cell types. Central to these processes is the modification and/or remodeling of chromatin by multisubunit complexes through association of site-specific DNA-binding proteins. Our laboratory and others have described a multisubunit complex in Drosophila, the Myb–Muv B (MMB) or dREAM complex (Korenjak et al. 2004; Lewis et al. 2004), which contains the previously identified five-subunit Myb complex (containing Myb, Caf1/p55, Mip40, Mip120, and Mip130) (Beall et al. 2002), in addition to E2f2, Rbf1 or Rbf2, DP, and Lin-52. Curiously, the complex contains both activator (Myb) and repressor (Mip130, Rbf1, Rbf2, and E2f2/DP) proteins. This point and that MMB is widely expressed in different tissues led to the idea that MMB may function both as an activator or repressor at specific chromosomal locations. Depending on the developmental pathways in particular tissues or cell types, different signals might regulate switching of function at a particular site. We suggested that at sites known to be repressed by MMB, Myb is a silent member not participating in the transcriptional repression, even though Myb itself is present at the cis-acting site (Korenjak et al. 2004; Lewis et al. 2004), and that activation of MMB at a subsequent time would depend on Myb function.
Our finding that Lethal (3) Malignant Brain Tumor [L(3)MBT], NURF, and the histone deacetylase Rpd3 associate with MMB suggests that histone binding and/or modification are possible mechanisms by which MMB acts to repress transcription and/or replication (Lewis et al. 2004). Thus, MMB and individual subunits are poised to change their measured role by switching off repression (or activation) in a given cell lineage by post-transcriptional modifications or association of coactivators (or repressors).
The genes regulated by MMB in Drosophila tissue culture cells are primarily differentiation and development-specific genes, and most often, MMB is a transcriptional repressor (Dimova et al. 2003; Lewis et al. 2004). Recent genomic profiling in Kc cells of five MMB members (Mip130, Mip120, Myb, E2F2, and Lin-52) showed that these proteins were bound together at thousands of chromosome sites, and RNA interference (RNAi) experiments revealed that MMB participated in either transcriptional repression or activation for many genes (D. Georlette, S. Ahn, P. Lewis, D. MacAlpine, E. Cheung, E. Beall, J. Manak, and M. Botchan, in prep.). In cell culture and in vivo, the accumulation of Myb and E2F2 proteins, but not mRNAs, depends on the integrity of MMB: Loss of Mip130 dramatically affects the levels of both proteins (Beall et al. 2004; Korenjak et al. 2004). These data, together with the biochemical finding that essentially all of Myb is found in complex with MMB, led to the proposal that most if not all of the phenotypes previously identified as Myb-specific (or E2F2-specific) must be evaluated in terms of loss of MMB function in either myb or e2f2 mutants.
myb is an essential gene in Drosophila (Katzen et al. 1985; Manak et al. 2002). However, mutations in the largest subunit of MMB, mip130, are viable and suppress myb lethality (Beall et al. 2004). Furthermore, homozygous mip130 mutant females have drastically reduced fecundity. Cytological and developmental studies of egg chambers from several MMB subunit mutants were critical in building a heuristic model for MMB function. Normally in the ovary, a developmentally controlled replication program occurs in the somatic follicle cell nuclei surrounding the developing oocyte. In these nuclei, overall genomic replication ceases at stage 9 during egg chamber development and is followed at stage 10 by specific DNA replication at four loci that results in amplification of the genes critical for egg shell formation (Orr-Weaver et al. 1989; Calvi et al. 1998; Claycomb et al. 2004). Myb binds to the well-defined enhancer for one such amplicon in vivo. When myb is removed by genetic manipulation, replication no longer occurs at the four foci, demonstrating a direct and positive role for Myb in replication at these sites (Beall et al. 2002). In contrast, mip130 mutant ovaries display global genomic replication in amplification-stage follicle cell nuclei, indicating a negative role for mip130 in replication at sites other than at the chorion origins (Beall et al. 2004). Based on these observations and the data summarized above, we previously suggested that MMB functions as either an activator or repressor of chromosomal functions depending on the chromosomal and developmental context. Critically in this model, the essential function of Myb is to activate a repressive complex to which it belongs. In its absence, this unchecked repressive activity by the partial MMB complex is lethal. Our presumption was that animals lacking MMB (as in mip130 mutants) maintained expression (or repression) of normal target genes through less robust or redundant mechanisms, resulting in viability of these mutants. Furthermore, a critical, but previously untested prediction of this model is that an MMB complex devoid of Myb could still be targeted to chromosomes.
In order to gain further insight into the role(s) for MMB in vivo, we generated a mutation in the second largest subunit, mip120. In addition, we identified a P-element-induced allele in another subunit, mip40. As with mip130, we found that mip40 and mip120 mutants were viable and displayed either sterility (mip120) or reduced fecundity (mip40) of mutant females. Moreover, mip40 and mip120 suppressed myb lethality, again suggesting that the essential function of Myb in vivo is to counter a repressive activity of MMB. Immunostaining of polytene chromosomes revealed that the association of Myb with specific chromosomal sites was dependent on Mip120, reinforcing the idea that Myb needs MMB for chromatin binding. Conversely, Mip120 and Mip130 did not require Myb for polytene chromosome binding.
Unanticipated and in contrast to mip130 mutants, we found that mip40 and mip120 mutant males were sterile, thus defining a new role for these proteins in male fertility. Given that Mip130 has a paralog in the testis called Aly (White-Cooper et al. 2000), we investigated the possibility that Mip40 and/or Mip120 might either function in a testis-specific Aly complex or that one or both might have testis-specific paralogs.
We used a combination of affinity, ion-exchange, and gel filtration chromatography to isolate Mip40-containing complexes from testis nuclear extracts. In addition to MMB, we identified tMAC, a new testis-specific meiotic arrest complex in which the only MMB subunits found were Mip40 and Caf1/p55, in addition to the testis-specific proteins Aly, Cookie monster (Comr), Matotopetli (Topi), and Tomb. Our model is that MMB functions as a cell-type- and developmental-stage-specific regulator of transcription and replication with various subunits contributing to a “Swiss Army Knife” type of versatility—the ability to interact specifically with numerous cis-elements, and to interact with numerous coactivators or corepressors as determined by context. This versatility extends beyond MMB itself, as some subunits are part of other tissue-specific complexes involved in gene expression.
Results
mip120 and mip40 mutants are viable
Searches of the existing mutant databases revealed that there were P-element insertions available within or nearby both mip40 and mip120. The line EY16520 contained a P-element insertion into the coding sequence of mip40 (181 base pairs [bp] from the ATG), which we designated mip40EY16520 (Fig. 1A), and was homozygous viable (Table 1). Line KG05422 contained a P-element insertion into the first intron of mip120 and did not disrupt the coding sequence (Fig. 1B). Complicating the genetic analysis, another gene, CG6050 or eftu-M, was also contained within the first intron of mip120.
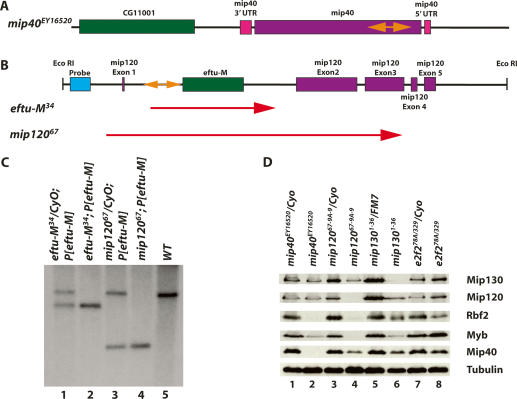
Figure 1.
Molecular verification of the mip40 and mip120 mutants. (A) The chromosomal configuration of the mip40 locus is shown (purple and pink), along with the location of the P-element insert in the mip40-coding sequence (orange double-sided arrow). (B) The chromosomal configuration of the mip120 locus is shown, with the mip120 exons in pink. The location of eftu-M (CG6050) within the first mip120 intron is shown in green. The P-element insertion for line KG05422 is shown in orange. The location of the probe used in C (blue) and the EcoRI restriction sites are also shown. The two lines generated after mobilization of the P-element and used in this study, eftu-M34 and mip12067, are shown. The regions deleted in these mutants are indicated by the red arrows. (C) Southern blot of eftu-M34 and mip12067 containing a transgene expressing eftu-M on the third chromosome (P[eftu-M]). Shown are EcoRI digests of DNA isolated from the strains indicated on the top and probed with the DNA fragment shown in B. The size of the deletion in each mutant as determined by sequence analysis is as follows: eftu-M34, 2236 bp; mip12067, 5770 bp. (D) Whole animals of the indicated genotypes were homogenized in sample buffer. “e2f2” refers to flies derived from crossing e2f2329/CyoKrGFP and e2f278A/CyoKrGFP (Cayirlioglu et al. 2001). Fly equivalents (0.3) were loaded onto a 9% SDS-PAGE, immunoblotted, and analyzed for the accumulation of the indicated MMB subunit using affinity-purified antibodies. Tubulin served as the loading control.
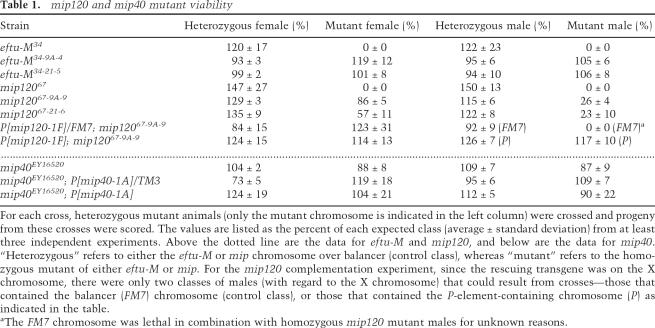
Table 1.
mip120 and mip40 mutant viability
For each cross, heterozygous mutant animals (only the mutant chromosome is indicated in the left column) were crossed and progeny from these crosses were scored. The values are listed as the percent of each expected class (average ± standard deviation) from at least three independent experiments. Above the dotted line are the data for eftu-M and mip120, and below are the data for mip40. “Heterozygous” refers to either the eftu-M or mip chromosome over balancer (control class), whereas “mutant” refers to the homozygous mutant of either eftu-M or mip. For the mip120 complementation experiment, since the rescuing transgene was on the X chromosome, there were only two classes of males (with regard to the X chromosome) that could result from crosses—those that contained the balancer (FM7) chromosome (control class), or those that contained the P-element-containing chromosome (P) as indicated in the table.
aThe FM7 chromosome was lethal in combination with homozygous mip120 mutant males for unknown reasons.
In order to generate mutants in mip120, we performed a P-element excision screen using line KG05422 (for details, see Materials and Methods). From 130 excision lines generated, 17 were homozygous lethal. None of the homozygous viable lines contained deletions of mip120 protein-coding sequence (data not shown). Of the lethal lines analyzed, four had deletions of eftu-M only (including line 34, or eftu-M34), and one (line 67, or mip12067) had a deletion of both eftu-M and most of mip120 (Fig. 1B).
eftu-M34 mutant chromosomes were homozygous lethal, indicating that eftu-M might be an essential gene. Therefore, the lethality observed with mip12067 may or may not be solely due to deletion of eftu-M. In order to resolve this issue, eftu-M-containing transgenic lines were generated and tested for their ability to complement loss of eftu-M in both mutants. Third-chromosome transgenic lines were initially tested for complementation, because mip120 was on the second chromosome. A transgene, P[eftu-M # 17], fully complemented eftu-M34 mutant lethality (data not shown). When P[eftu-M # 17] was placed into mip12067 mutants, homozygous mutant viability was now observed, demonstrating that mip120 was not an essential gene. Therefore, the lethality observed in mip12067 mutants was solely due to deletion of eftu-M. Southern blot analysis demonstrated the size of the deletions in each line (Fig. 1C), and sequencing of regions flanking the deletion determined the exact break points (Fig. 1B).
In order to simplify further analyses, we placed eftu-M-containing transgenes onto the chromosomes harboring either the eftu-M34 or mip12067 mutations. We created recombinants from two different second chromosome P[eftu-M]-containing transgenic lines (9A and 21) and both eftu-M34 and mip12067 (to generate eftu-M34-9A-4, eftu-M34-21-5, mip12067-9A-9, and mip12067-21-6). As expected, we found that P[eftu-M] completely complemented the lethality of eftu-M34 mutants (Table 1, eftu-M34-9A-4 and eftu-M34-21-5). We also found that mip12067-9A-9 and mip12067-21-6 were homozygous viable (Table 1). Moreover, eftu-M34-9A-4 and eftu-M34-21-5 were fertile and were easily maintained as a stock (data not shown). Therefore, we analyzed phenotypes of mip12067 mutants containing P[eftu-M] transgenes, provisionally concluding that all the phenotypes ascribed to these mutants were due to loss of mip120 function.
myb; mip40 and myb; mip120 double mutants are viable
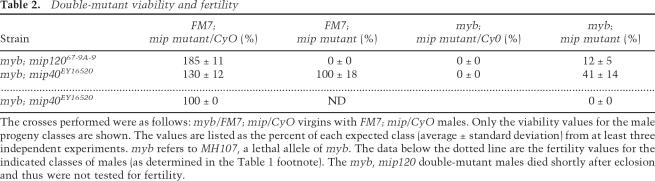
Because myb; mip130 double-mutant flies are viable (Beall et al. 2004), we postulated that mutations in additional MMB subunits (such as mip40 and mip120) that remove the repressive activities of MMB would similarly suppress the lethality of myb mutant animals. We found that both mybMH107; mip40EY16520 and mybMH107; mip12067-9A-9 double mutants were viable (Table 2). PCR using primers specific to each allele confirmed the double-mutant viability in each case (Supplementary Fig. 1). As with mip40EY16520 mutant males alone (see below), we found that mybMH107; mip40EY16520 double-mutant males were sterile (Table 2). Unfortunately, the mybMH107; mip12067-9A-9 mutant males were so unhealthy that they died soon after eclosing and were unable to be tested for fertility. The viability of myb; mip120 and myb; mip40 double mutants therefore suggests that mip130, mip40, and mip120 are all epistatic to myb.
Table 2.
Double-mutant viability and fertility
The crosses performed were as follows: myb/FM7; mip/CyO virgins with FM7; mip/CyO males. Only the viability values for the male progeny classes are shown. The values are listed as the percent of each expected class (average ± standard deviation) from at least three independent experiments. myb refers to MH107, a lethal allele of myb. The data below the dotted line are the fertility values for the indicated classes of males (as determined in the Table 1 footnote). The myb, mip120 double-mutant males died shortly after eclosion and thus were not tested for fertility.
Myb accumulation and chromatin association are compromised in mip mutants
We asked if the protein levels of five of the MMB members were compromised in mip40, mip120, mip130, and e2f2 mutants. While loss of E2f2 had no effect on Myb accumulation, we found that Myb levels were severely reduced or absent when any of the Mips were removed (Fig. 1D). In fact, there was little to no effect upon removal of E2f2 for any MMB member examined, except for a small decrease in the amount of Rbf2. Unexpectedly, we found that Rbf2 accumulation was severely reduced in mip40EY16520 and mip12067-9A-9 mutants. Given that the major binding partner for Rbf2 was thought to be E2f2 (Stevaux et al. 2002), it was not anticipated that Rbf2 accumulation was critically dependent on Mip40 and Mip120 and only marginally dependent on E2f2. The reduced protein levels were not due to effects on transcription for each protein examined since transcript levels were not affected in each mutant as determined by RT–PCR analysis (Supplementary Fig. 2). In fact, we found that the most dramatic effect upon MMB member accumulation occurred when either Mip120 or Mip130 was removed, suggesting that these two subunits are “core” components critical for maintaining the integrity of MMB.
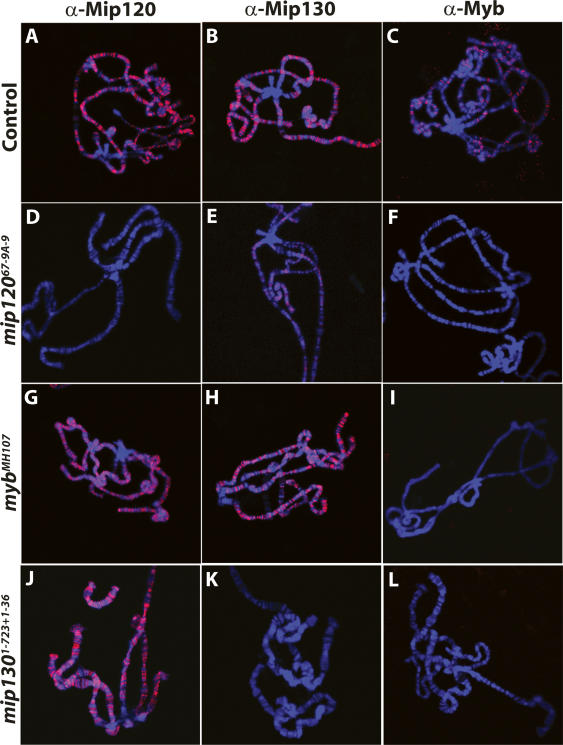
In these and previous studies, loss of core MMB proteins led to a dramatic loss in the total levels of Myb protein. Nevertheless, a reduced or absent signal by immunoblot analysis still leaves open the possibility that proteins may still be present and bound to DNA. Although we found that there is not a significant pool of free MMB complex or individual subunits (data not shown), we wanted to address this issue directly. We looked at the staining pattern of Mip120, Mip130, and Myb on polytene chromosomes in control, mip120, mip130, and myb mutants. We found that all three proteins were bound to many sites throughout all five chromosome arms in control spreads, with a conspicuous absence of staining at the heterochromatic chromocenter (Fig. 2A–C). This lack of centric heterochromatic Mip120 staining in the control spreads is different from the results previously reported (Korenjak et al. 2004) and may be due to different staining protocols. The Myb signal was absent in mip12067-9A-9 and mip1301–723 + 1–36 mutants (Fig. 2F,L), strongly suggesting that Myb was critically dependent on MMB complex for chromosome association. In addition, we found that Mip120 and Mip130 were still localized to many sites in mybMH107 mutants (Fig. 2G,H), consistent with our hypothesis that the inability to activate chromatin-bound MMB complexes lacking Myb is the explanation for mybMH107 mutant lethality (Beall et al. 2004).
Figure 2.
Chromatin association of Myb is dependent on Mip120 and Mip130. Confocal images of polytene chromosome spreads using GFP-sorted wandering third instar larvae from the following strains are shown: mip12067-9A-9/CyOKrGFP, mip1301–723 + 1–36/FM7KrGFP or mybMH107/FM7cAcGFP. The control (GFP+) or mutant (GFP−) spreads are as indicated on the left. (Blue) DAPI; (red) Anti-Mip120, Anti-Mip130, or Anti-Myb as indicated on the top.
We found that loss of Mip120 had the greatest overall effect upon chromatin binding of individual MMB subunits. In addition to the complete loss of Myb and Mip120 signal, we found a severe reduction in Mip130 signal in mip12067-9A-9 mutants (Fig. 2D–F). Given that there were reduced amounts of Mip130 protein in mip12067-9A-9 mutants (Fig. 1D, lane 4) and that Mip130 contains AT-hook motifs capable of binding to A/T-rich DNA, it is not surprising that small amounts of Mip130 were still detected on chromatin in mip12067-9A-9 mutants. Conversely, Mip120 protein was still present in mip1301–723 + 1–36 mutants (Fig. 1D, lane 6). As Mip120 can bind to DNA on its own (Beall et al. 2002), it was not surprising that Mip120 was found throughout the genome in mip1301–723 + 1–36 mutants (Fig. 2J).
All together, the immunoblot and polytene immunostaining data support our model that members of MMB are dependent on each other for stability as part of a large multisubunit complex that is bound to many specific chromosomal sites. As all of our antibodies were raised in rabbits, we could not colocalize different proteins simultaneously to see if MMB members were bound to the same sites throughout the whole genome. Moreover, epitope masking of subunits caused by different coactivators or corepressors at specific loci may result in obscuring some of the actual locations of individual subunits. However, we found a striking similarity in the banding pattern for Myb, Mip120, and Mip130 on the small fourth chromosome, where the binding pattern of MMB is relatively simple (Supplementary Fig. 3).
mip120 mutants have eye abnormalities
Examination of the phenotypes for both mip40 and mip120 mutants revealed both similarities and differences. Contrary to mip40EY16520 mutants, mip12067; P[eftu-M # 17] mutants were generally unhealthy and appeared smaller than nonmutant siblings, and males died within a day or so of eclosion. Although not systematically studied, mip12067; P[eftu-M # 17] females died sporadically within 2–10 d after eclosion.
A striking mip12067; P[eftu-M # 17] mutant phenotype was that the animals displayed moderate to severe eye phenotypes including small or completely absent eyes and a rough appearance (Supplementary Fig. 4). Identical results were observed with mip12067-9A-9 (data not shown). In a small fraction of individuals whose eyes were examined, the eyes contained ommatidia patches completely isolated away from the major eye mass on some flies (Supplementary Fig. 4E). There was no difference in BrdU incorporation in control versus mip12067-9A-9 mutant eye disks (Supplementary Fig. 5), suggesting that the eye phenotype was not due to a gross replication problem during development. However, we cannot address subtle replication problems in mip12067-9A-9 mutant eye disks that were not detected by this method; for instance, in the number and location of origins that initiated DNA replication. Reasonably, the eye defects resulted in part because of altered gene expression patterns in ommatidia precursors that may have required Mip120 for either repression or activation of critical promoters.
mip40 and mip120 mutant females are sterile
We found that both mip40 and mip120 mutant females were sterile (Table 3). mip40EY16520 mutant sterility resembled that which we have previously reported for mip130 mutant females: Most individual mutant females tested were completely sterile, whereas some mutant females had one to a few larvae crawling in vials after 7 d of egg laying. This was in contrast to nearly every control sister tested that had upward of 100 larvae per vial. mip120 mutant females were more severely affected and never laid a single egg.
Table 3.
Summary of mip120 and mip40 mutant fertility
The descriptions of the crosses and classes of mutants are in the footnote for Table 1. At least five independent experiments, for a total of 50–100 animals, were performed for each genotype listed, and the values listed represent the average percent fertility ± standard deviation. Fertility was determined by scoring for the presence of at least one larva in each vial after 7 d at 25°C. (ND) Not done. The data for mip120 are above the dotted line, and the data for mip40 are below the dotted line.
aThe actual number of larvae per vial was ∼0.5%–1% of wild type.
In order to ensure that the mutant phenotypes that we were studying were due to loss of either mip40 or mip120 only, we performed complementation experiments using transgenic animals carrying either a mip40 genomic DNA fragment, or the mip120 cDNA under the control of its endogenous promoter (for details, see the Supplemental Material). We found that a mip40 transgene completely restored both the viability (Table 1) and fertility (Table 3) of mip40EY16520 mutants to levels comparable to control heterozygous mutant sisters. Similarly, we found that a mip120 transgene completely restored the viability (Table 1), fertility (Table 3), eye morphology, and normal longevity (data not shown) of mip12067-9A-9/67-21-6 mutants to those of control siblings.
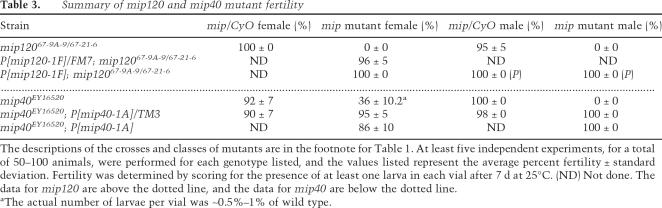
When we examined the BrdU incorporation and Orc2 localization pattern in ovaries dissected from both mip40 and mip120 mutant females, we observed very different things. Beginning in stage 10 egg chambers, there are normally four foci of BrdU incorporation and Orc2 localization in the follicle cell nuclei surrounding the developing oocyte that result from amplification of the DNA only at these sites (Fig. 3A,E). Many nuclei within a single mip40EY16520 mutant egg chamber displayed a BrdU incorporation pattern identical to wild type. However, dispersed throughout the same egg chamber were nuclei with BrdU incorporation and Orc2 staining that was spread throughout the entire nucleus (Fig. 3B). These data showed that Mip40 has negative regulatory roles in replication in amplifying follicle cell nuclei, similar to what was observed with mip130 mutants (Beall et al. 2004).
Figure 3.
Mip40 and Mip120 are required for egg chamber development. (A,B,E,F) Shown are merged confocal images of stage 10 egg chambers from females of the genotypes indicated on the bottom of each panel. (Red) Anti-Orc2; (green) Anti-BrdU; (blue) DAPI. (C,D) Same as in other panels, except that a lower-magnification view (10×) is shown to demonstrate the lack of stage 10 and later egg chambers in mip120 mutants.
In contrast, mip12067-9A-9/67-21-6 mutants had a complete lack of amplification-stage egg chambers (stage 10–14) (Fig. 3, cf. C and D). Even after examining ovaries from the very few long-lived surviving females obtained (∼7–10 d after eclosion), we never observed egg chambers that developed beyond stage 9. In the stages that could be examined, there was qualitatively less DNA replication in the follicle and nurse cell nuclei, based on the intensity and number of BrdU-positive nuclei. When we examined ovaries from mip12067-9A-9/67-21-6 mutants containing either one or two copies of the mip120-containing transgene, we found that all egg chamber stages were clearly apparent, including amplification stages. Moreover, the replication pattern and Orc2 localization were identical to controls, further demonstrating that the mip120-containing transgene completely complemented the mip12067-9A-9/67-21-6 mutant phenotypes (Fig. 3E,F). These data establish a role for Mip120 in ovary development prior to DNA amplification, where Myb, Mip130, and Mip40 are critical for normal egg shell production.
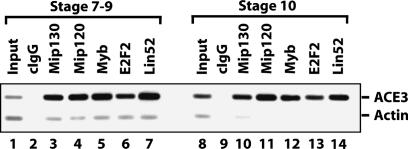
Previously, we established that members of MMB (Myb and Mip120) were bound to the third chromosome chorion locus, ACE3, in amplification-stage egg chambers as measured by chromatin immunoprecipitation (ChIP) (Beall et al. 2002). However, an important question that is germane to the idea that MMB may act as a functional switch at a particular chromosomal location in various developmental pathways arises: Is MMB bound to ACE3 prior to origin activation? In order to answer this question, we mass-isolated egg chambers at different stages (for details, see the Supplemental Material) and performed ChIP analysis focusing on ACE3 occupancy prior to amplification (stage 7–9) and at amplification-stage egg chambers (stage 10) (Fig. 4). We found that the five assayed members of MMB were, in fact, bound to ACE3 prior to and during amplification. These data are consistent with a switching model for MMB because both the positive-acting factors (like Myb) and the negative-acting factors (like E2F2) are associated with chorion origins as a complex in preamplification egg chambers and seem poised for action during amplification.
Figure 4.
The MMB complex is present at ACE3 prior to, and during, chorion gene amplification. Sheared cross-linked chromatin from preamplification (less than or equal to stage 9) and amplification (stage 10) egg chambers was used for ChIP using the indicated antibodies. Shown is an ethidium bromide-stained gel of PCR reactions performed using both ACE3-specific and actin-specific (control) primers simultaneously in the reaction containing either input or immunoprecipitated DNA. ACE3 is enriched for all samples except the control IgG and input samples. Note that the ratio of ACE3:actin is higher in the input DNA for amplification-stage egg chambers (lane 8) when compared with preamplification egg chambers (lane 1) due to higher copy numbers of the ACE3 locus in this sample.
mip40 and mip120 mutant males are sterile
Our previous studies with mip130 mutants showed that mutant males are fertile (Beall et al. 2004). In contrast, we found that both mip40 and mip120 mutant males were sterile. Male fertility for either mip40EY16520 or mip12067-9A-9/67-21-6 was restored to wild type with either a mip40- or mip120-encoding transgene, respectively (Table 3).
During normal testis development, germline stem cells divide to both maintain the stem cell population and produce spermatogonial cells. The spermatogonia undergo four mitotic cell divisions before differentiating to primary spermatocytes. These primary spermatocytes undergo high levels of transcription in preparation for terminal differentiation (for review, see Fuller 1998). Male sterile mutants have been identified at many different points during spermatogenesis, including a group of genes that, when mutated, cause arrest at the primary spermatocyte stage of development (Lin et al. 1996). These genes are called the meiotic arrest genes because mutants arrest at the G2–M transition of meiosis I, and cells contain partially condensed chromosomes that eventually accumulate and fill the testis. Meiotic arrest genes are required for both meiotic cell cycle progression and the onset of spermatid differentiation in mature spermatocytes. The currently identified meiotic arrest genes have been subdivided into two classes. The aly class genes, including aly and comr, regulate transcription of some genes required for entry into meiosis (boule, twine, Cyclin B) in addition to many spermiogenesis genes required for the differentiation of functional sperm (fuzzy onions, janus B, don juan, gonadal). In contrast, the can class genes (including cannonball [can], meiosis 1 arrest [mia], and spermatocyte arrest [sa]) do not affect transcription of the meiotic cell cycle genes but are required for spermiogenesis gene transcription (White-Cooper et al. 1998).
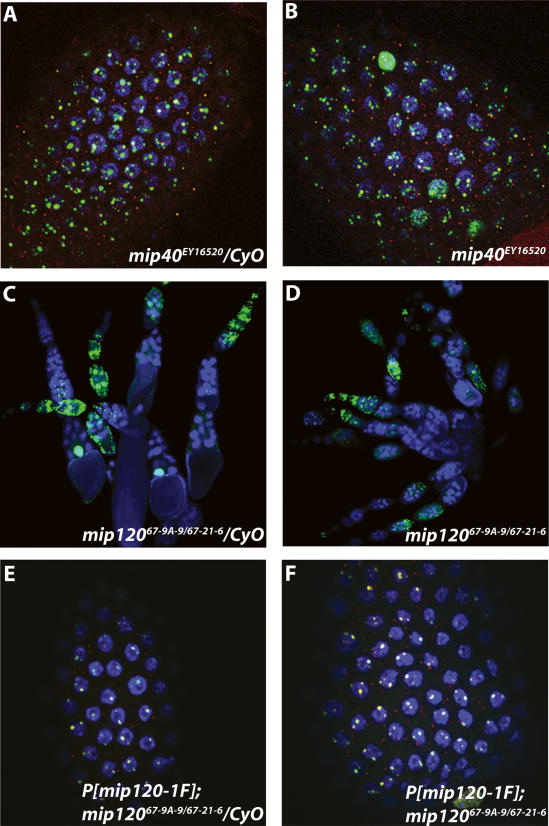
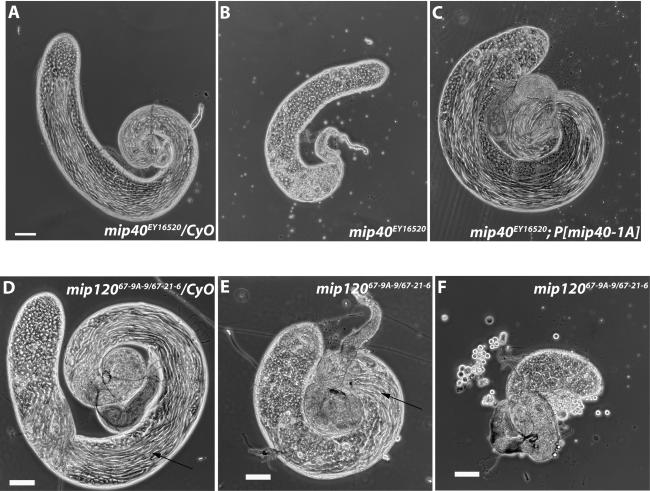
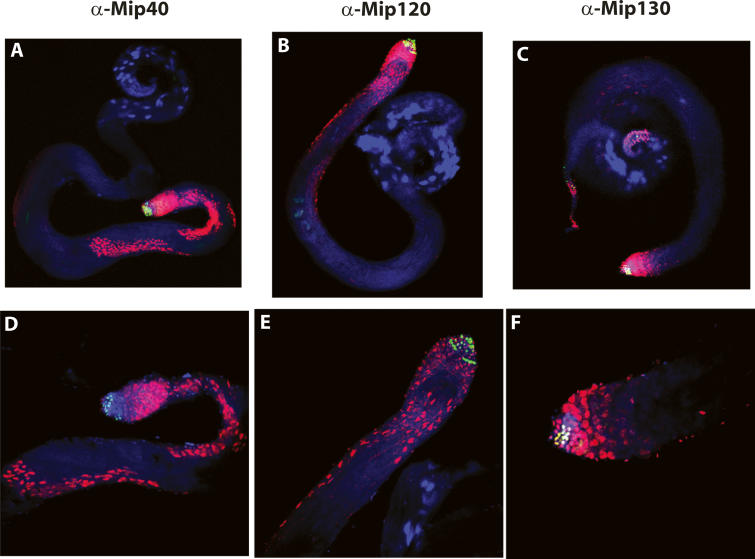
Phase-contrast microscopy revealed that testes from both mip40 and mip120 mutants had clear defects in development when compared with control siblings. mip40EY16520 mutants appeared to be arrested at the primary spermatocyte stage of development (Fig. 5A,B) and were complemented by a mip40-encoding transgene (Fig. 5C). Hoechst DNA staining suggested that the spermatocytes were arrested prior to the G2–M transition, similar to the arrest point for other meiotic arrest mutants (Supplementary Fig. 6). Meiotic cell cycle genes, but not spermiogenesis genes, were transcribed in testes from mip40 mutants, placing mip40 into the can class of meiotic arrest mutants (Supplementary Fig. 7). Also, Mip40 protein was detected in spermatogonia undergoing mitotic divisions: The highest Mip40 protein staining was found in spermatocytes (Fig. 6A,C), consistent with a role for Mip40 in spermatocyte development.
Figure 5.
Mip40 and Mip120 are required for male fertility. Shown are phase-contrast images using a Leica DM5000 microscope of testes dissected from mip40 (A–C) or mip120 (D–F) males of the genotypes indicated in each panel. (B) Note the complete lack of mature sperm in mip40EY16520 mutants due to presumed arrest at the spermatocyte stage of spermatid differentiation. Arrows identify sperm bundles present in control (D) and mip120 mutant (E) testes.
Figure 6.
Mip40 and Mip120 proteins are expressed in spermatocytes. Shown are merged confocal images of testes from wild-type males stained with the antibodies indicated on the top (red). (Green) Anti-BrdU; (blue) DAPI. (A–C) View (10×) of whole testis to show staining in the mitotic (green) and spermatocyte stages. (D–E) Higher-magnification image (25×) to show detail.
In contrast, mip120 mutant testes appeared very different from mip40 mutants, and the phenotype varied from male to male. All mip12067-9A-9/67-21-6 testes were smaller in appearance than control siblings and appeared to contain some germ cell stages up to and beyond the spermatocyte stage, including sperm (Fig. 5, cf. E,F and D). When dissected, the sperm from mip12067-9A-9/67-21-6 mutant testes were completely immotile. In contrast to Mip40 staining, we found that the highest Mip120 staining levels were in the earliest stages of spermatogonial development, with some expression apparent in spermatocyte stages (Fig. 6B,E). Interestingly, Mip130 protein staining was strongest in the earlier spermatogonial stages as well, with even lower expression in the spermatocyte stages (Fig. 6C,F). The different mip mutant male fertility phenotypes are consistent with the different protein expression profiles in testes and significantly separate physiological functions of the MMB genes. mip40 appears to be required for meiosis and spermatid differentiation independent of mip120 and mip130. The biochemical data presented below show that Mip40 is part of another complex that we suggest executes function(s) at the spermatocyte stage of testis development.
Mip40 is a subunit in a testis-specific complex required for spermatid differentiation
The Mip130 family member Aly is thought to transcriptionally activate genes involved in both spermatid differentiation and the meiotic cell cycle in primary spermatocytes (Jiang and White-Cooper 2003; Perezgasga et al. 2004). aly mutant males are sterile, and testes from aly mutants contain cells arrested at the spermatocyte stage that fail to undergo meiosis to produce sperm—similar to what was observed in testes from mip40EY16520 males. These facts coupled with the distinct mip40 mutant phenotypes when compared with other MMB subunit mutants fostered a simple idea: Perhaps Mip40 functions intimately with non-MMB factors, such as Aly, in the testis to carry out the meiotic cell cycle and spermatid differentiation transcriptional program(s). Conceptually, Mip40 could either work through MMB by direct or indirect association with Aly or independent of MMB in another unanticipated way.
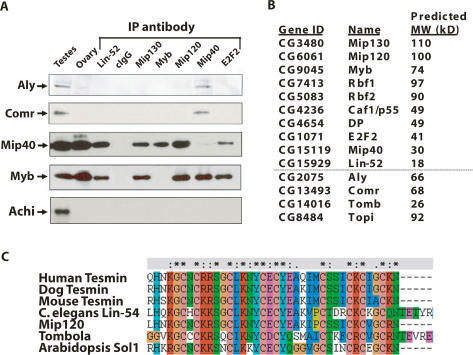
To test this idea, we mass-isolated testes from adults and prepared nuclear extracts (for details, see the Supplemental Material). As estimated by light microscopy, the testes comprised ∼50%–60% of the total mass in the sieved material from which the nuclear extracts were made. We fractionated extracts using Poros heparin chromatography, pooled the Aly-containing fractions, and performed immunoprecipitations (IPs) to determine whether Mip40 and other MMB subunits associated with Aly. We found that of the MMB complex members tested, only Mip40 coimmunoprecipitated both Aly and a known interacting partner of Aly, Comr (Fig. 7A). Because ethidium bromide was included in the IPs, the interactions were not mediated through DNA.
Figure 7.
Meiotic arrest proteins specifically interact with Mip40. (A) Poros-heparin fractions of Drosophila testis extracts were immunoprecipitated using the antibodies indicated at the top. The precipitates were washed with buffer containing 0.5 M KCl, and the coimmunoprecipitated proteins were eluted with 0.4% sarcosyl. As a result of the sarcosyl wash, the protein to which the antibody was raised will remain in the pellet and will not appear in the IP supernatant. All immunoprecipitations were performed in the presence of 50 μg/mL ethidium bromide. Shown are the immunoblot analyses of dissected whole testes, ovaries, and the immunoprecipitated material using the antibodies indicated on the left. Only Mip40 antibodies coimmunoprecipitated Aly and Comr specifically. It also appears that MMB is an abundant complex relative to tMAC, based on the qualitative signal strengths of immunoblot signals for Aly and Comr versus Mip40 and Myb in the testis lane. (B) Data from duplicate control and Mip40 antibody columns were analyzed by mass spectroscopy. Above the dotted line is a list of MMB subunits identified by mass spectrometry from the Mip40 antibody column eluate that were not present in the control antibody column eluate. In addition, several testis-specific proteins that have known functions in meiotic arrest were also identified only in the Mip40 eluate, including Aly, Comr, Tomb, and Topi. (C) Drosophila Tomb belongs to the Tesmin/TSO family of proteins. Shown is an alignment of the CXC domain of Tomb and other tesmin-family proteins: Human Tesmin (AAH64579), Dog Tesmin (XP_854573), Mouse Tesmin (BAB64935), Drosophila Mip120, Drosophila Tombola, and Arabidopsis SOL1. (*) Conserved residues; (:) similar residues among the aligned proteins.
Aly and Comr have been reported to interact with a pair of homeobox transcription factors of the TGIF subclass, Achi and Vis, in Drosophila testis extracts. Achi and Vis are essential for spermatogenesis and are implicated in transcriptional activation along with Aly and Comr of several testis-specific transcripts (Wang and Mann 2003; Perezgasga et al. 2004). We failed to detect Achi by immunoblot analysis of the coimmunoprecipitated proteins from MMB subunits (Fig. 7A), and Achi or Vis peptides were not detected by mass spectrometry in the Mip40 affinity column (Fig. 7B). The absence of Achi and Vis in our IPs suggested that there might be more than one pool of Aly and Comr proteins in spermatogonia, where Aly/Comr form a complex with Achi/Vis in the absence of Mip40. Alternatively, the reported interaction between Achi/Vis and Aly/Comr could have been mediated by contaminating DNA in the coimmunoprecipitations.
In order to look for additional protein(s) that might be associated with Mip40, Aly, and Comr, we applied the Poros heparin fractions onto an anti-Mip40 affinity column and used mass spectrometry to identify interacting proteins in the pooled eluate. As a control, a mock purification was performed using unspecific total rabbit IgG. Because the MMB complex was also purified on this column, we found that all MMB complex members were present as anticipated (Fig. 7B). Among the additional proteins specifically associated with Mip40, we found Aly, Comr, Topi, and Tomb. Topi is a testis-specific zinc-finger transcription factor that is also a meiotic arrest member in which mutants arrest at the spermatocyte stage of development (Perezgasga et al. 2004). Tomb is a Mip120 family member that contains significant homology with Mip120 in the cysteine-rich domain that is conserved from nematodes to humans (Fig. 7C). Included in this family are the testis-specific Tesmin proteins from humans, mice, and dogs. Tomb is significantly smaller than Mip120 (26 kDa vs. 100 kDa) and contains a single cysteine-rich domain (whereas the other family members all contain two) that shares significant homology with Mip120. The Caenorhabditis elegans Mip120 homolog, Lin-54, binds to DNA and interacts with other synMuv class B proteins (Deplancke et al. 2006; Harrison et al. 2006). Moreover, another family member, CPP1, binds to DNA through this cysteine-rich domain (Cvitanich et al. 2000), and it is possible that all family members use this domain to contact DNA directly.
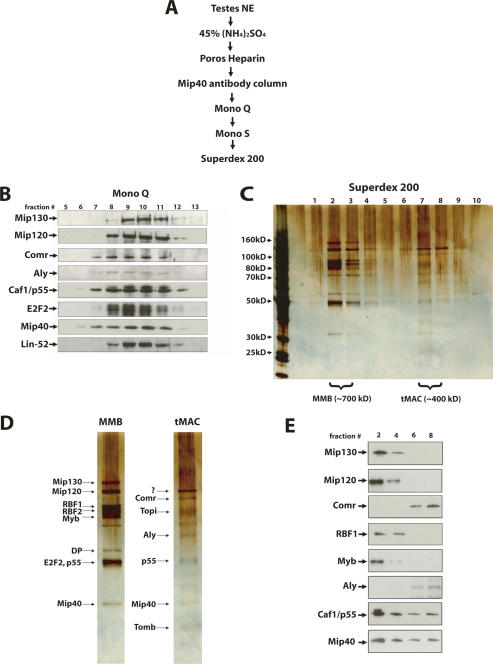
Intrigued by the mass spectrometry data, we wondered whether a complex distinct from MMB could be isolated from testes that contained Mip40 and the testis-specific proteins identified thus far by IP. We fractionated testis nuclear extracts using a combination of anti-Mip40 affinity, ion-exchange, and gel filtration chromatography as described in Figure 8A. We found that MMB and the Aly-associated factors cofractionated on a Mono Q column (Fig. 8B), but were readily separated on a final Superdex PG200 gel filtration column (Fig. 8C). On this column, MMB eluted with an apparent molecular weight of 700 kDa, whereas the Aly complex that clearly contained Mip40 eluted with an apparent molecular weight of 400 kDa. Based on known and predicted molecular weights of factors identified by mass spectrometry, we assigned names to the bands visualized by silver stain for the peak Superdex 200 fractions (Fig. 8D). There are several additional undetermined bands in the Mip40–Aly complex, most strikingly, a prominent band above Comr that is not Mip120. Further work will be needed to identify these proteins. To confirm the identity of the bands that were assigned, we performed immunoblot analysis of Superdex PG200 fractions using antibodies that were currently available to us (Fig. 8E). We found that the only MMB complex members that were part of the distinct Mip40–Aly complex were Mip40 and Caf1/p55, and there was no overlap between other members of MMB and the newly identified complex. Together, these data define a new, testis-specific complex that contains members of MMB. Based on the developmental arrest phenotype of mutant alleles for the genes encoding many of the founding members of the complex, we named this new complex tMAC.
Figure 8.
Mip40 and Aly are present in a newly identified complex in testes. (A) The fractionation scheme used to purify tMAC. Fractions from each chromatography step were immunoblotted for Mip40, Comr, and Aly. Peak fractions were pooled and used for the subsequent chromatography step. (B) Aly and Comr polypeptides cofractionated with MMB using ion-exchange chromatography. Shown are immunoblots of the Mono Q fractions indicated on the top of the panel using antibodies directed against Aly, Comr, and subunits of the MMB complex as indicated on the left. (C) Fractions eluting from the final Superdex 200 gel filtration column were resolved on a 4%–12% gradient SDS-PAGE and visualized by silver staining. The peaks of MMB (fractions 2,3) and tMAC (fractions 7,8) are indicated at the bottom. The MMB and tMA complexes eluted with an estimated molecular weight of ∼700 kDa and ∼400 kDa, respectively. Molecular weight markers are indicated on the left. (D) The peaks of MMB (fraction 2) and tMAC (fraction 7) from C are compared. Proteins identified by size and confirmed by immunoblot analysis (see below) are indicated on the left of each panel. Bands labeled as Topi and Tomb are based on the predicted molecular weight of the full-length polypeptides. The band marked with a “?” represents an unknown cofractionating protein that is not Mip120. (E) The only MMB proteins present in tMAC are Mip40 and Caf1/p55. Shown are immunoblots of the Superdex 200 gel filtration fractions indicated at the top using antibodies against the MMB complex subunits Aly and Comr as indicated on the left.
Discussion
Many homologous genes encoding MMB subunits were first discovered in nematodes and defined the synMuv B (synthetic multivulva) pathway. Extensive genetic analysis has established pleiotropic functions of these genes in C. elegans (e.g., RNAi effects, suppression of multivulva, G1–S transition, germline effects) (Boxem and van den Heuvel 2002; Wang et al. 2005; Ceol et al. 2006; Chi and Reinke 2006), and interestingly, many of the factors found in MMB are also in a multiprotein complex in worms (Harrison et al. 2006). The primary goal of our work was to elucidate the functions of the Drosophila MMB complex in vivo. To that end, we created a mutation in the second largest subunit of MMB, mip120, and characterized a P-element insertion line in another subunit, mip40. In addition to observing phenotypes in common to those already described for other members of MMB (such as female sterility), we discovered new mutant phenotypes that reveal roles for the MMB members Mip40 and Mip120 during development and differentiation of diverse cell types. These include eye development for mip120 and testes development for both mip40 and mip120.
These genetic findings that clearly separate developmental functions for Mip40 and Mip120 do not provide mechanistic insights into how the pleiotropic effects are manifested. For example, partial MMB complexes, resulting from loss-of-function alleles, may assume neomorphic activity. We have previously argued that myb lethality in Drosophila is a consequence of such effects. The model that rationalizes “silent subunits” present at a given location to promote switching from repression to activation (or vice versa) adds genetic complexity to the timing of critical execution functions for different MMB factors. This model anticipates that loss-of-function alleles of genes for different MMB subunits would manifest different arrest points. We also considered the possibility that MMB subunits do not always function as a unit. We were curious about the finding that mip40 mutant males displayed a staining pattern for Mip40 protein in testes quite distinct from Mip130 or Mip120. Epitope masking of one or another protein in MMB, due to changing coactivator or corepressor association, might explain such staining patterns. However, further work led to the search for a putative testis-specific complex that contained Mip40, where it reasonably would act in a distinct way from its functions in MMB.
tMAC: a testis-specific paralog of MMB
We purified a complex from testis nuclear extracts that contains MMB members Mip40 and Caf1/p55, in addition to the testis-specific meiotic arrest proteins Aly, Comr, Topi, and Tomb. We named this complex tMAC, because aly, comr, topi, and mip40 mutants all display the same testis phenotype: an arrest at the primary spermatocyte stage of development, consistent with the notion that they are all acting together in a complex to promote differentiation and meiotic cell cycle progression. It seems likely that other proteins might interact with tMAC to aid in the regulation of testis-specific transcripts. This idea stems from what we already know about MMB, where proteins such as Rpd3 and L(3)MBT physically associate with MMB only during early steps in the biochemical purification process and are critical for function at different DNA sites. To date, we have not been successful in isolating any other alternative or subcomplexes containing members of MMB from either embryo or tissue culture nuclear extracts. Furthermore, genomic profiling in KC cells of key MMB members (Myb, E2F2, Lin-52, Mip120, and Mip130) substantiates the hypothesis that these proteins work together as a group rather than as isolated factors on DNA (D. Georlette, S. Ahn, P. Lewis, D. MacAlpine, E. Cheung, E. Beall, J. Manak, and M. Botchan, in prep.). Had we not begun with testis-enriched starting material, it would have been impossible for us to identify tMAC. Thus, despite the present data showing that the MMB core factors always work as an ensemble, it is possible that some of the pleiotropic phenotypes observed for different MMB subunit mutants could reflect the activity of a subunit functioning outside of MMB.
It is striking that both tMAC and MMB contain proteins, other than Mip40 and Caf/p55, that are similar to each other in domain architecture: Aly (tMAC) or Mip130 (MMB) and Tomb (tMAC) or Mip120 (MMB). Given that MMB and tMAC contain multiple site-specific DNA-binding proteins (Myb, E2F2/DP, Mip120, Mip130 in MMB; and Tomb and Topi in tMAC), a potentially large number of genes may be regulated by tMAC as we now know is true for MMB.
Antisera against the MMB subunit, Lin-52, failed to coimmunoprecipitate Comr or Aly; therefore, it is not likely a tMAC subunit. However, it is interesting to note that there is another Lin-52 family member in Drosophila (CG12442) (see Supplementary Fig. 8A for alignment). The adult Drosophila gene expression database (http://flyatlas.org) indicates that this gene is, indeed, highly expressed in testis relative to other tissues. Given that this protein is extremely small (predicted molecular weight of 16 kDa), it is possible that it was not present in sufficient quantities to be detected in the mass spectrometry analysis and may in fact be part of tMAC. If so, that would be the third MMB subunit to have an alternative “testis-specific” form present in tMAC.
Gonad-specific forms of proteins that are ubiquitously expressed and generally found in complexes that regulate transcription may, indeed, be a common theme. For example, gonad-specific components of the basal RNA polymerase II transcription machinery are crucial for developmentally regulated gene expression programs in these tissues (Hochheimer and Tjian 2003). Five testis-specific TATA-binding protein-associated factors (TAFs) have been identified in Drosophila (encoded by the can, sa, mia, nht, and rye genes) (Hiller et al. 2001, 2004). All are required in spermatocytes for the normal transcription of target genes involved in post-meiotic spermatid differentiation (the so-called can class of genes) (White-Cooper et al. 1998). It is thought that these testis-specific TAFs may associate with some of the general TAF subunits to create a testis-specific TFIID (tTFIID) that carries out the developmentally regulated transcriptional program in spermatocytes. The mip40-null allele is also in the can class, suggesting that tMAC may interact with tTFIID at can class gene promoters. It will be interesting to explore the possibility that tMAC is a testis-specific coactivator with tTFIID. Other tMAC subunits fall into the aly class and might be what is expected for a large complex paralogous to MMB, where one or another subunit may be silent and subsequently required at a later stage or developmental pathway.
Mip130 family members, such as Aly, share a domain that is called a “DIRP” (domain in Rb-related pathway) domain that is thought to be responsible for interaction with Rb. The DIRP domain of human-Lin-9 (Mip130) is necessary for association with Rb; however, the interaction between hLin-9 and Rb may be indirect as hLin-9 may exist in a complex with other proteins that directly touch Rb (Gagrica et al. 2004). We did not detect either of the two Drosophila Rb proteins in tMAC-containing fractions. When alignments were made with Mip130 family members, we noticed a region within the DIRP domain that was conserved between all family members except Aly. It is possible that this divergent region within the DIRP domain is critical for Rb interaction in other family members and has been lost in Aly (Supplementary Fig. 8B). Although we have no direct understanding of how Aly works for transcriptional activation, it is possible that tMAC contains both activating and repressing components similar to MMB and that repression at particular loci does not require E2F/Rb.
Ovary phenotypes for mutant alleles of MMB-encoding genes
When examined for replication profiles in mutant ovaries, we found an absence of amplification-stage egg chambers in mip120 mutants, and widespread BrdU incorporation and Orc2 staining in mip40 mutant amplification-stage follicle cell nuclei. The mip40 egg chamber phenotype is similar to that of mip130 and is consistent with a negative regulatory role for these proteins in genome-wide replication at these stages. We suggest that both Mip40 and Mip120 are functioning in complex with MMB in ovaries and that both are required for normal patterns of replication in amplification-stage follicle cell nuclei. We speculate, based in part on our unpublished studies of the intricate regulatory network of MMB in Kc cells, that the different mutant phenotypes may simply reflect differences in gene expression profiles that result when individual MMB complex members are missing. A key role for Mip120 in the stability of chromatin-bound MMB (Fig. 2) might, therefore, explain the more severe phenotype of mip120 mutants. More specifically, MMB may regulate the expression of genes critical for amplification-stage egg chamber development either directly or indirectly, and Mip120 is required for targeting MMB to these gene promoters at a particular developmental stage prior to amplification stages. In contrast to Mip120, Myb, and E2F2/DP, Mip40 has no direct DNA-binding ability. Mip40 may be required for repression or activation only after MMB is targeted to chromosomal sites.
As with myb; mip130 mutants, we found that myb; mip40 and myb; mip120 double mutants were viable, further demonstrating that function(s) of MMB without Myb are responsible for myb lethality. Myb protein was no longer associated with chromatin in mip120 and mip130 mutant polytene spreads. However, staining of polytene chromosomes demonstrated that Mip120 and Mip130 proteins were still bound to chromatin in myb mutants in such a way that may prove lethal in the absence of myb. Together, these data support a model in which Myb is critically dependent on members of the MMB complex for both stability and association with chromatin.
We suggest that the presence of MMB at the replication enhancer ACE3 in stage 7–9 egg chambers (Fig. 4) may actively repress DNA replication here and at other sites in the genome. MMB at ACE3 at these early stages seems poised to await signals for initiation of amplification. The conversion of a repressive MMB complex into an activating complex may require cyclin E/Cdk2 activity, which is required for amplification (Calvi et al. 1998). In this context it is likely that Rbf association with MMB will persist during amplification as Rbf association with MMB remains unchanged even after saturating hyperphosphorylation by Cdk:cyclin E in vitro (Lewis et al. 2004). In the future, determining the cell-type-specific signals that target MMB at well-defined cis-regulatory elements at both the follicle cell amplicons and in other tissues will help unravel how MMB functions in vivo.
Materials and methods
Genetics
See the Supplemental Material for details about strains, P-element moblilization screen, complementation analysis, and myb double-mutant analysis.
Antibody staining and BrdU labeling
Ovaries or testes were processed and stained as described (Beall et al. 2004). See the Supplemental Material for details.
Polytene chromosome immunostaining
Mutant mip120, mip130, or myb wandering third instar larvae were sorted from GFP-balancer controls and polytene chromosome spreads prepared and stained using standard methods. See the Supplemental Material for details.
Purification of staged Drosophila egg chambers and testes
Canton S flies (50,000–100,000) were gently pulse-blended and applied to a column of stainless-steel mesh seives as suggested in Ashburner (1989). A Drosophila Laboratory Manual. A detailed protocol is described in the Supplemental Material.
ChIP
Staged egg chambers were formaldehyde-fixed, sheared chromatin was prepared, and immunoprecipitations were performed as described in detail in the Supplemental Material.
DNA staining of unfixed testis
Testes were dissected and placed into 30 μL of Hoechst staining solution (1 mg/mL Hoechst 33342 in dH2O, diluted to 1:500–1:1000 in PBS) prior to squashing. Testes were observed by phase-contrast and epifluorescence microscopy on a Leica DM5000 upright microscope. Images were captured using Leica’s FireCam software.
RNA in situ hybridization
Whole-mount RNA in situ hybridization was performed using standard methods. Ribonucleotide probes were generated from linearized plasmids using the Roche Molecular Biochemicals RNA-labeling kit.
MMB and tMAC purifications and immunoprecipitations
Nuclear extracts from testes were prepared and chromatographed as described for embryos in Lewis et al. (2004) and in detail in the Supplemental Material.
Mass spectroscopy
Samples from Mip40 and cIgG column were TCA-precipitated and washed with −20°C acetone. The pellet was resuspend in 8 M urea, 100 mM Tris-Cl (pH 8.5) solution, 5 mM TCEP, and 10 mM iodoacetamide. The urea concentration was diluted to 2 M with Tris (pH 8.5) followed by the addition of 0.5 μg sequencing grade trypsin, and was incubated overnight at 37°C. Three-dimensional analysis was performed by the Proteomics/Mass Spectrometry Facility (University of California at Berkeley).
Acknowledgments
We thank Todd Laverty for help with the polytene spreads, Lori Kohlstaedt for help with the mass spectroscopy, Helen White-Cooper for anti-Aly and anti-Comr antibodies, and Zhaohui Wang for anti-Achi antibodies. This work was supported by NIH grant R39 CA-30490 to M.R.B.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1516607
References
- Ashburner M. Drosophila: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Beall E.L., Manak J.R., Zhou S., Bell M., Lipsick J.S., Botchan M.R., Manak J.R., Zhou S., Bell M., Lipsick J.S., Botchan M.R., Zhou S., Bell M., Lipsick J.S., Botchan M.R., Bell M., Lipsick J.S., Botchan M.R., Lipsick J.S., Botchan M.R., Botchan M.R. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Beall E.L., Bell M., Georlette D., Botchan M.R., Bell M., Georlette D., Botchan M.R., Georlette D., Botchan M.R., Botchan M.R. Dm-myb mutant lethality in Drosophila is dependent upon mip130: Positive and negative regulation of DNA replication. Genes & Dev. 2004;18:1667–1680. doi: 10.1101/gad.1206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem M., van den Heuvel S., van den Heuvel S. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr. Biol. 2002;12:906–911. doi: 10.1016/s0960-9822(02)00844-8. [DOI] [PubMed] [Google Scholar]
- Calvi B.R., Lilly M.A., Spradling A.C., Lilly M.A., Spradling A.C., Spradling A.C. Cell cycle control of chorion gene amplification. Genes & Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Bonnette P.C., Dickson M.R., Duronio R.J., Bonnette P.C., Dickson M.R., Duronio R.J., Dickson M.R., Duronio R.J., Duronio R.J. Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development. 2001;128:5085–5098. doi: 10.1242/dev.128.24.5085. [DOI] [PubMed] [Google Scholar]
- Ceol C.J., Stegmeier F., Harrison M.M., Horvitz H.R., Stegmeier F., Harrison M.M., Horvitz H.R., Harrison M.M., Horvitz H.R., Horvitz H.R. Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development. Genetics. 2006;173:709–726. doi: 10.1534/genetics.106.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Reinke V., Reinke V. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (EF2) does not require LIN-35 (pRB) Development. 2006;133:3147–3157. doi: 10.1242/dev.02490. [DOI] [PubMed] [Google Scholar]
- Claycomb J.M., Benasutti M., Bosco G., Fenger D.D., Orr-Weaver T.L., Benasutti M., Bosco G., Fenger D.D., Orr-Weaver T.L., Bosco G., Fenger D.D., Orr-Weaver T.L., Fenger D.D., Orr-Weaver T.L., Orr-Weaver T.L. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev. Cell. 2004;6:145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- Cvitanich C., Pallisgaard N., Nielsen K.A., Hansen A.C., Larsen K., Pihakaski-Maunsbach K., Marcker K.A., Jensen E.O., Pallisgaard N., Nielsen K.A., Hansen A.C., Larsen K., Pihakaski-Maunsbach K., Marcker K.A., Jensen E.O., Nielsen K.A., Hansen A.C., Larsen K., Pihakaski-Maunsbach K., Marcker K.A., Jensen E.O., Hansen A.C., Larsen K., Pihakaski-Maunsbach K., Marcker K.A., Jensen E.O., Larsen K., Pihakaski-Maunsbach K., Marcker K.A., Jensen E.O., Pihakaski-Maunsbach K., Marcker K.A., Jensen E.O., Marcker K.A., Jensen E.O., Jensen E.O. CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proc. Natl. Acad. Sci. 2000;97:8163–8168. doi: 10.1073/pnas.090468497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Mukhopadhyay A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Mukhopadhyay A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Sequerra R., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Doucette-Stamm L., Reece-Hoyes J.S., Hope I.A., Reece-Hoyes J.S., Hope I.A., Hope I.A., et al. A gene-centered C. elegans protein–DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Dimova D.K., Stevaux O., Frolov M.V., Dyson N.J., Stevaux O., Frolov M.V., Dyson N.J., Frolov M.V., Dyson N.J., Dyson N.J. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes & Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M.T. Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin. Cell Dev. Biol. 1998;9:433–444. doi: 10.1006/scdb.1998.0227. [DOI] [PubMed] [Google Scholar]
- Gagrica S., Hauser S., Kolfschoten I., Osterloh L., Agami R., Gaubatz S., Hauser S., Kolfschoten I., Osterloh L., Agami R., Gaubatz S., Kolfschoten I., Osterloh L., Agami R., Gaubatz S., Osterloh L., Agami R., Gaubatz S., Agami R., Gaubatz S., Gaubatz S. Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J. 2004;23:4627–4638. doi: 10.1038/sj.emboj.7600470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.M., Ceol C.J., Lu X., Horvitz H.R., Ceol C.J., Lu X., Horvitz H.R., Lu X., Horvitz H.R., Horvitz H.R. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. 2006;103:16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M.A., Lin T.Y., Wood C., Fuller M.T., Lin T.Y., Wood C., Fuller M.T., Wood C., Fuller M.T., Fuller M.T. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes & Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M., Chen X., Pringle M.J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Chen X., Pringle M.J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Pringle M.J., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Suchorolski M., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Sancak Y., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Viswanathan S., Bolival B., Lin T.Y., Marino S., Fuller M.T., Bolival B., Lin T.Y., Marino S., Fuller M.T., Lin T.Y., Marino S., Fuller M.T., Marino S., Fuller M.T., Fuller M.T. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Hochheimer A., Tjian R., Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes & Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Jiang J., White-Cooper H., White-Cooper H. Transcriptional activation in Drosophila spermatogenesis involves the mutually dependent function of aly and a novel meiotic arrest gene cookie monster. Development. 2003;130:563–573. doi: 10.1242/dev.00246. [DOI] [PubMed] [Google Scholar]
- Jiang J., Benson E., Bausek N., Doggett K., White-Cooper H., Benson E., Bausek N., Doggett K., White-Cooper H., Bausek N., Doggett K., White-Cooper H., Doggett K., White-Cooper H., White-Cooper H. Tombola, a tesmin/TSO1-family protein, regulates tanscriptional activation in the Drosophila male germline and physically interacts with Always early. Development. 2007 doi: 10.1242/dev.000521. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen A.L., Kornberg T.B., Bishop J.M., Kornberg T.B., Bishop J.M., Bishop J.M. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell. 1985;41:449–456. doi: 10.1016/s0092-8674(85)80018-0. [DOI] [PubMed] [Google Scholar]
- Korenjak M., Taylor-Harding B., Binne U.K., Satterlee J.S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A., Taylor-Harding B., Binne U.K., Satterlee J.S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A., Binne U.K., Satterlee J.S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A., Satterlee J.S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A., Aasland R., White-Cooper H., Dyson N., Brehm A., White-Cooper H., Dyson N., Brehm A., Dyson N., Brehm A., Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Lewis P.W., Beall E.L., Fleischer T.C., Georlette D., Link A.J., Botchan M.R., Beall E.L., Fleischer T.C., Georlette D., Link A.J., Botchan M.R., Fleischer T.C., Georlette D., Link A.J., Botchan M.R., Georlette D., Link A.J., Botchan M.R., Link A.J., Botchan M.R., Botchan M.R. Identification of a Drosophila Myb–E2F2/RBF transcriptional repressor complex. Genes & Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.Y., Viswanathan S., Wood C., Wilson P.G., Wolf N., Fuller M.T., Viswanathan S., Wood C., Wilson P.G., Wolf N., Fuller M.T., Wood C., Wilson P.G., Wolf N., Fuller M.T., Wilson P.G., Wolf N., Fuller M.T., Wolf N., Fuller M.T., Fuller M.T. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development. 1996;122:1331–1341. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Mitiku N., Lipsick J.S., Mitiku N., Lipsick J.S., Lipsick J.S. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc. Natl. Acad. Sci. 2002;99:7438–7443. doi: 10.1073/pnas.122231599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Johnston C.G., Spradling A.C., Johnston C.G., Spradling A.C., Spradling A.C. The role of ACE3 in Drosophila chorion gene amplification. EMBO J. 1989;8:4153–4162. doi: 10.1002/j.1460-2075.1989.tb08600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perezgasga L., Jiang J., Bolival B., Jr., Hiller M., Benson E., Fuller M.T., White-Cooper H., Jiang J., Bolival B., Jr., Hiller M., Benson E., Fuller M.T., White-Cooper H., Bolival B., Jr., Hiller M., Benson E., Fuller M.T., White-Cooper H., Hiller M., Benson E., Fuller M.T., White-Cooper H., Benson E., Fuller M.T., White-Cooper H., Fuller M.T., White-Cooper H., White-Cooper H. Regulation of transcription of meiotic cell cycle and terminal differentiation genes by the testis-specific Zn-finger protein matotopetli. Development. 2004;131:1691–1702. doi: 10.1242/dev.01032. [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dimova D., Frolov M.V., Taylor-Harding B., Morris E., Dyson N., Dimova D., Frolov M.V., Taylor-Harding B., Morris E., Dyson N., Frolov M.V., Taylor-Harding B., Morris E., Dyson N., Taylor-Harding B., Morris E., Dyson N., Morris E., Dyson N., Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Mann R.S., Mann R.S. Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development. 2003;130:2853–2865. doi: 10.1242/dev.00510. [DOI] [PubMed] [Google Scholar]
- Wang D., Kennedy S., Conte D., Jr., Kim J.K., Gabel H.W., Kamath R.S., Mello C.C., Ruvkun G., Kennedy S., Conte D., Jr., Kim J.K., Gabel H.W., Kamath R.S., Mello C.C., Ruvkun G., Conte D., Jr., Kim J.K., Gabel H.W., Kamath R.S., Mello C.C., Ruvkun G., Kim J.K., Gabel H.W., Kamath R.S., Mello C.C., Ruvkun G., Gabel H.W., Kamath R.S., Mello C.C., Ruvkun G., Kamath R.S., Mello C.C., Ruvkun G., Mello C.C., Ruvkun G., Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- White-Cooper H., Schafer M.A., Alphey L.S., Fuller M.T., Schafer M.A., Alphey L.S., Fuller M.T., Alphey L.S., Fuller M.T., Fuller M.T. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- White-Cooper H., Leroy D., MacQueen A., Fuller M.T., Leroy D., MacQueen A., Fuller M.T., MacQueen A., Fuller M.T., Fuller M.T. Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development. 2000;127:5463–5473. doi: 10.1242/dev.127.24.5463. [DOI] [PubMed] [Google Scholar]