Abstract
Systematic genome-wide studies to map genomic regions associated with human diseases are becoming more practical. Increasingly, efforts will be focused on the identification of the specific functional variants responsible for the disease. The challenges of identifying causal variants include the need for complete ascertainment of genetic variants and the need to consider the possibility of multiple causal alleles. We recently reported that risk of systemic lupus erythematosus (SLE) is strongly associated with a common SNP in IFN regulatory factor 5 (IRF5), and that this variant altered spicing in a way that might provide a functional explanation for the reproducible association to SLE risk. Here, by resequencing and genotyping in patients with SLE, we find evidence for three functional alleles of IRF5: the previously described exon 1B splice site variant, a 30-bp in-frame insertion/deletion variant of exon 6 that alters a proline-, glutamic acid-, serine- and threonine-rich domain region, and a variant in a conserved polyA+ signal sequence that alters the length of the 3′ UTR and stability of IRF5 mRNAs. Haplotypes of these three variants define at least three distinct levels of risk to SLE. Understanding how combinations of variants influence IRF5 function may offer etiological and therapeutic insights in SLE; more generally, IRF5 and SLE illustrates how multiple common variants of the same gene can together influence risk of common disease.
Keywords: interferon pathway, systemic lupus erythematosus
The number of loci convincingly associated with complex diseases has risen in recent years because of increasing knowledge of the human genome and its variation, cost-effective high-throughput genotyping technologies, and improved methods for statistical analysis. The identification of alleles reproducibly associated with disease has the potential to define genes and biological pathways as playing a causal role in vivo in the population. Recent illustrations include the role of the complement pathway in age-related macular degeneration (1–3), the IL-23 pathway in inflammatory bowel disease (4), and IFN regulatory factor 5 (IRF5) in systemic lupus erythematosus (SLE) (5, 6). For example, the type I IFN pathway is dysregulated in SLE, with many IFN-inducible genes overexpressed in the peripheral blood of SLE patients; the finding of robust association to SLE of SNPs in IRF5, a transcription factor downstream of the type I IFN and Toll-like receptors (7–10), provided the first direct and specific evidence that variation in the type I IFN pathway plays a causal role in SLE pathogenesis (5, 6).
Although there is increasing progress in identifying loci convincingly associated with complex disease, in few cases have the causal mutations responsible for the association unambiguously been identified. Several factors make this difficult: (i) for efficiency, initial screens survey only a subset of human variants to identify a region; (ii) because of linkage disequilibrium, multiple variants in each region may show equivalent signals of association; (iii) causal variants may be noncoding, and there is no analogue to the genetic code to identify from primary sequence data alleles impacting gene expression, regulation, or posttranslational modifications; and (iv) there may be multiple causal variants at a locus, with prominent examples including multiple HLA alleles in autoimmune diseases (11), multiple alleles of complement factor H in age-related macular degeneration (1–3), and multiple alleles at the APOE locus influencing lipids and Alzheimer's disease (12, 13).
These general challenges are illustrated by the association of common variants in IRF5 to risk of SLE. We recently demonstrated that common alleles of IRF5 are robustly associated with risk of SLE in both family- and population-based cohorts (5, 6). The marker most associated with risk to SLE (rs2004640) was found to alter a consensus splice donor site and to allow expression of isoforms bearing exon 1B (an alternative exon 1). This combination of genetic and functional data provided a potential model to explain the effect of IRF5 on SLE (5). However, as described below, a more extensive assessment of common variation in IRF5 in SLE provides evidence for three statistically independent signals of association to SLE, one of which is stronger than that of the exon 1B splice site (rs2004640).
Results
Characterization of Sequence Variation at IRF5.
To more fully characterize genetic variation at IRF5, we sequenced the exons and 1 kb upstream of the IRF5 exon 1A in DNA from 136 cases of SLE; and we sequenced each of the introns in 40 SLE cases and 8 controls [supporting information (SI) Tables 3 and 4]. In total, 52 variants were observed, of which 26 were previously identified (present in dbSNP), and 32 were undescribed. Of the variants not in the database, 13 had minor allele frequency >1%. Each such variant was genotyped in the HapMap Centre d'Etudes du Polymorphisme Humain (CEPH) Utah residents with ancestry from northern and western Europe (CEU) samples, allowing them to be integrated with data from the International HapMap Project.
Although no common single-nucleotide missense variants of IRF5 were observed, a 30-bp in-frame insertion/deletion (indel) in exon 6 was observed. The exon 6 indel is located in a proline-, glutamic acid-, serine- and threonine-rich domain, a motif previously shown to influence protein stability and function in the IRF family of proteins (14). TagSNPs were selected to serve as proxies (r2 > 0.8) for all SNPs with minor allele frequency >1% in the combined data from HapMap Phase II (15) and genotype data in the same samples for the SNPs discovered in our sequencing effort.
Association of Common Variation in IRF5 to Risk of SLE.
Each tagSNP was individually tested for association to SLE in a combined trio and family collection of 555 families from the U.S. and the United Kingdom (Table 1). The strongest association with SLE was for three highly correlated SNPs (rs2070197, rs10488631, and rs12539741, pairwise r2 >0.95). These SNPs (which we refer to as Group 1) do not include the previously studied exon 1B splice site variant and showed highly significant association: Transmitted/Untransmitted (T/U) ratio = 1.8; P = 1.2 × 10−7. To assess whether the Group 1 variants could explain the association to SLE, we performed conditional logistic regression incorporating one of the Group 1 SNPs (rs2070197). This model was rejected, because a second set of correlated SNPs (referred to as Group 2, rs729302, rs4728142, rs2004640, and rs6966125) were independently associated with risk to SLE (P < 0.002–0.008, Table 1; see SI Table 5). Group 2 includes the previously studied exon 1B splice site variant (rs2004640).
Table 1.
Single-marker transmission and conditional analyses in SLE trios from the United States and United Kingdom
| Marker | Allele | T* | U | P† | P conditional on Group 1 variants (rs2070197) | P conditional on Group 1 (rs2070197) and Group 2 (rs2004640) variants | P conditional on Group 1 (rs2070197), Group 2 (rs2004640), and Group 3 (rs10954213) variants |
|---|---|---|---|---|---|---|---|
| rs1495461 | G | 260 | 220 | 0.068 | 0.37 | 0.59 | 0.45 |
| rs960633 | T | 257 | 209 | 0.026 | 0.54 | 0.75 | 0.56 |
| rs729302 | A | 270 | 195 | 5.0 × 10-4 | 0.0024 | 0.56 | 0.82 |
| rs4728142 | A | 363 | 257 | 2.1 × 10−5 | 0.0054 | 0.0096 | 0.20 |
| rs3807135 | C | 298 | 241 | 0.0141 | 0.28 | 0.0008 | 0.31 |
| rs2004640 | T | 344 | 233 | 3.8 × 10−6 | 0.0019 | — | — |
| rs752637 | G | 297 | 238 | 0.011 | 0.28 | 0.0010 | 0.14 |
| Exon 6 indel | A | 337 | 294 | 0.087 | 0.25 | 0.01 | NA |
| rs2070197 | C | 205 | 111 | 1.2 × 10−7 | — | — | — |
| rs10954213 | A | 282 | 226 | 0.013 | 0.14 | 0.0089 | — |
| rs11770589 | G | 338 | 288 | 0.046 | 0.16 | 0.01 | NA |
| rs10954214 | T | 281 | 232 | 0.031 | 0.14 | 0.02 | NA |
| rs10488631 | C | 223 | 125 | 1.5 × 10−7 | 1.00 | NA | NA |
| rs2280714 | A | 268 | 219 | 0.026 | 0.18 | 0.0078 | NA |
| rs12539741 | T | 222 | 125 | 1.9 × 10−7 | 1.00 | NA | NA |
| rs17166351 | C | 336 | 290 | 0.066 | 0.17 | 0.005 | NA |
| rs696612 | C | 153 | 124 | 0.081 | 0.0078 | 0.825 | 0.29 |
*Number of transmitted (T) and untransmitted alleles (U).
†Nominal P value for association to SLE and P value for the association to SLE, under the model that the indicated markers fully explain the association to SLE, as determined by conditional logisitic regression. NA indicates that the association to SLE cannot be calculated because it is statistically indistinguishable from the proposed model. Only SNPs with a P value <0.1 are listed in the table.
To test the hypothesis that the combination of Group 1 and Group 2 variants fully account for the association observed to SLE, we repeated the conditional logistic regression analysis including a Group 1 and a Group 2 variant in the model (represented by rs2070197 and rs2004640). A third set of six highly correlated SNPs (referred to as Group 3; rs4728142, rs3807135, rs752637, rs10954213, rs2280714, and rs17166351) were associated with risk of SLE (P < 0.001–0.01, Table 1; see SI Table 5).
These results indicate that three independent sets of correlated IRF5 variants (Groups 1, 2, and 3) each provide statistically independent evidence for association with risk of SLE. In previous limited surveys of genetic variation (5, 6), the exon 1B splice site (rs2004640) was most strongly associated with SLE and, given its potential functional role in splicing, offered a potential explanation for how IRF5 variation might influence risk. After a more extensive assessment of IRF5 variation, however, it is clear that rs2004640 is no longer adequate to explain all of the effect of IRF5 on risk to SLE; indeed, it is not even the strongest contributor. We set out to identify other putative functional alleles that might explain the independent signals of association observed for Groups 1 and 3.
Cis-Acting Alleles Underlying Variation in IRF5 Expression.
One approach to finding causal alleles is to examine other phenotypes that might be less complex in their inheritance, providing power to distinguish the effects of highly correlated alleles, and to offer in vitro assays to assess function. In vitro expression levels provide one such phenotype (16, 17). Given our previous observation that one of the Group 3 variants (rs2280714) is associated with levels of IRF5 mRNA expression, we systematically examined the more complete set of IRF5 variants for alleles that might be associated to levels of IRF5 mRNA expression in lymphoblastoid cell lines.
The same set of tagSNPs genotyped in the SLE family cohort were studied in the HapMap samples (15), allowing correlation of genotype to mRNA expression data collected at the Sanger Institute (www.sanger.ac.uk/humgen/genevar). A variant in the 3′ UTR (rs10954213, Group 3) showed the strongest association with IRF5 expression: P = 3.5 × 10−55 (SI Table 6). This variant and one other (rs10954214) reside in conserved elements within the 3′ UTR, a region that often contains sequences that influence mRNA expression (18).
To increase power to distinguish effects of correlated SNPs, we genotyped a subset of the associated IRF5 variants in an independent data set of 233 CEPH samples for which microarray gene expression data were publicly available (17) (SI Table 7). Again, rs10954213 was the best predictor of IRF5 expression. Specifically, rs10954213 showed stronger associations than either the neighboring rs10954214 or the SNP we had studied previously, rs2280714 (SI Tables 7 and 8 and Fig. 1a). Formally, rs10954213 remained strongly associated with IRF5 mRNA levels after conditioning on rs2280714 (P = 5 × 10−9), whereas conditioning on rs10954213 nearly eliminates association of rs2280714 to IRF5 expression (P = 0.004). Finally, similar findings were observed for expression of IRF5 in whole blood of SLE cases (SI Fig. 3).
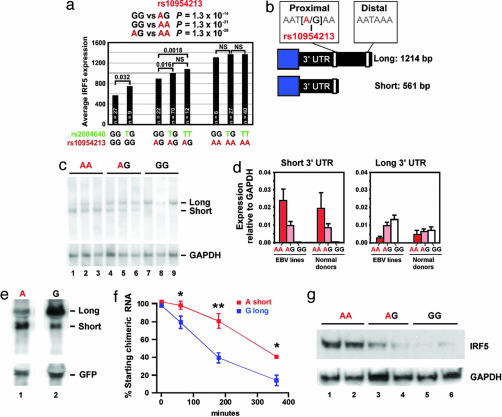
Fig. 1.
Expression levels of IRF5 mRNA are influenced by a polymorphism in a proximal 3′ UTR polyA+ signal sequence. (a) Microarray data from 233 CEU cell lines carrying various genotypes for rs2004640 and rs10954213 were examined for expression levels of IRF5. (b) Schematic of the 3′ UTR region of IRF5. (c) Northern blot of 500 ng of polyA+ RNA from cell lines carrying the indicated genotypes at rs10954213 (three cell lines from unrelated individuals for each genotype) using a common proximal 3′ UTR probe. Blots were stripped and reprobed for GAPDH. (d) Quantitative TaqMan RT-PCR in EBV cell lines (n = 9) and in control peripheral blood mononuclear cells (n = 14) for levels of IRF5 isoforms carrying the short or long 3′ UTR. (e) Northern blot using β-globin and GFP cDNA probes in Tet-off 293 cells transfected with chimeric β-globin:IRF5 3′ UTR expression plasmids for either the A or G allele of rs10954213. GFP expression plasmids were cotransfected as a control. (f) Graph shows the decay of β-globin:3′ IRF5 UTR mRNAs after suppression of new transcription with doxycycline. Results represent four independent experiments. ∗, P < 0.05; ∗∗, P < 0.01. (g) Western blot for IRF5 in two cell lines for each of the indicated genotypes at rs10954213 by using monoclonal anti-IRF5 antibodies that recognize a C-terminal peptide sequence of IRF5. Blots were stripped and reprobed with antibodies to GAPDH. Representative of five independent experiments.
These results indicate that rs10954213 is the best predictor of IRF5 expression in our survey of lymphoblastoid cell lines, clearly distinguishable in its effect from the other SNPs with which it is in strong linkage disequilibrium. Because rs10954213 is also a member of Group 3, it became a candidate for explaining the association of Group 3 SNPs to SLE (above). We note that the greater strength of the signal of association of IRF5 expression levels (P < 10−55) allowed us to distinguish the signal of rs10954213 from the other members of Group 3 for IRF5 expression, whereas we were not able to clearly distinguish the weaker signals of association to risk of SLE.
Although rs10954213 is the strongest determinant of IRF5 expression in our survey of common variation at IRF5, conditioning on this SNP did not account for all of the variance in IRF5 expression. After conditioning on rs10954213, the exon 1B splice site (rs2004640) and other linked SNPs were the next strongest association to IRF5 levels (SI Table 7). Specifically, the presence of the T allele at rs2004640, which allows the expression of exon 1B isoforms, was associated with significantly higher levels of IRF5 expression in cell lines carrying GG or AG genotype at rs10954213 (Fig. 1a). After incorporating a two-locus model of both rs10954213 and rs2004640, no other SNP has a nominally significant association to IRF5 expression in the CEU cell lines (SI Tables 7 and 9).
Thus, a systematic search for common variation that influences levels of IRF5 mRNA identified rs10954213, a SNP in a conserved an element within the 3′ UTR and a member of Group 3, as well as the exon 1B splice site variant (rs2004640), a member of Group 2.
A Group 3 Variant Alters a Polyadenylation Signal and Influences IRF5 Expression.
Although we previously had shown that the exon 1B SNP influences IRF5 mRNA levels through its effect on splicing (5), the function, if any, of rs10954213 was unknown. We observed that the sequence surrounding rs10954213 has been highly conserved throughout evolution (SI Fig. 4). Moreover, the rs10954213 G allele is predicted to disrupt a polyA+ signal sequence (AAUAAA→AAUGAA) located 552 bp downstream of the stop codon of IRF5 in the 3′ UTR region of exon 9. The canonical motif is a binding site for a protein complex called cleavage and polyadenylation specificity factor (CPSF). During RNA polymerase II transcription, CPSF binds to the AAUAAA sequence and is part of a complex that cuts the mRNA strand 10–30 bp downstream of the polyA+ signal and initiates polyadenylation of the transcripts (19).
Based on the location of rs10954213 in a conserved CPSF site, we hypothesized that the different alleles of rs10954213 might influence polyadenylation and thereby the length and stability of the IRF5 message. Specifically, we hypothesized that the A allele of rs10954213 might allow efficient polyadenylation ≈12 bp downstream, whereas the G allele favors the use of a distal polyA+ site 648 bp downstream (Fig. 1b).
To directly test this hypothesis, we performed two types of experiments: Northern blotting and quantitative PCR of IRF5 mRNA from cell lines and peripheral blood mononuclear cells of known genotype at rs10954213 and creation of chimeric mRNAs that attach the two alleles of the 3′ UTR to heterologous expression constructs.
We isolated total and polyA+ enriched RNA from the HapMap CEU population, selecting individuals based on genotype at rs10954213. Northern blotting of polyA+ RNA showed that cell lines homozygous for the A allele at rs10954213, carrying the wild-type AAUAAA on both alleles, expressed mainly a short version of IRF5 mRNAs (Fig. 1c). In contrast, cell lines homozygous for the G allele (AAUGAA) expressed almost exclusively a longer mRNA that used the second downstream polyA+ site (Fig. 1c). AG heterozygote cell lines showed expression of both isoforms. Identical results were obtained in Northern blots of total RNA isolated from the cell lines (data not shown). We further confirmed these results with TaqMan quantitative PCR assays in both EBV-transformed cell lines and normal donor peripheral blood mononuclear cells (Fig. 1d). These data confirmed that the allele at rs10954213 determines the site of polyadenylation; we now refer to rs10954213 as the polyA+ variant, with the A allele termed the “short” allele and the G allele the “long” allele.
To determine whether the long allele of the 3′ UTR might be unstable, we cloned the two versions of the 3′ UTR downstream of the coding region of rabbit β-globin and transfected 293 Tet-off kidney cells with expression plasmids driving either the chimeric cDNAs carrying either the short or long allele. Northern blotting 48 h after transfection showed that chimeric cDNAs used the expected polyA+ site (Fig. 1e), and that the long mRNAs had a shorter half-life than short chimeric transcripts (Fig. 1f). Estimates for the half-life of these transcripts, based on regression curves, were: 342 ± 88 min for the short allele and 125 ± 21 min for the long allele. By comparison, the calculated half-life of β-globin mRNA alone (lacking the IRF5 3′ UTR) was 11,631 ± 1,574 min.
These experiments document that disruption of the proximal polyA+ signal by rs10954213 leads to the transcription of long and relatively unstable IRF5 mRNA transcripts. These effects on IRF5 mRNA are reflected in levels of IRF5 protein, as shown by Western blots of whole cell lysates from EBV cell lines carrying the various polyA+ SNP genotypes: cells carrying the AA genotype showed ≈5-fold higher levels of immunoreactive IRF5 protein than cells carrying the GG genotype (Fig. 1g).
Exon 6 Indel and Risk of SLE.
Our previously published results and those above suggest that (i) the association of Group 2 SNPs to SLE is likely explained by the exon 1B splice site allele (rs2004640), and (ii) the association of the Group 3 SNPs is likely because of the polyA+ variant (rs10954213). In contrast, we found that none of the Group 1 SNPs alter the coding region of IRF5, lie in evolutionarily conserved regions, or change an annotated sequence motif. Either the Group 1 SNPs (or an undiscovered but strongly linked mutation) have an as-yet-unrecognized effect on IRF5 function, or the Group 1 SNPs have no functional consequence but instead tag a combination of other functional variants in IRF5.
To assess the second model (having found no evidence for a functional allele among the Group 1 SNPs), we performed the conditional logistic regression analysis not in order of statistical significance (as above) but instead starting with the two putative functional alleles identified above (exon 1B splice site variant and polyA+ variant). We observed multiple variants that showed significant association to SLE in this analysis (SI Table 10) and including the 30-bp in-frame indel polymorphism that we had discovered within exon 6 (Fig. 2a). This indel is located in a proline-, glutamic acid-, serine- and threonine-rich domain known to influence protein stability and function in the IRF family of proteins. Previous studies have shown that IRF5 protein isoforms, which, in part, differ by the 30-bp (10-aa) exon 6 indel (which had previously been observed in cDNA but not recognized to be a germ-line polymorphism) have differential ability to initiate transcription of IRF5 target genes (8, 20, 21).
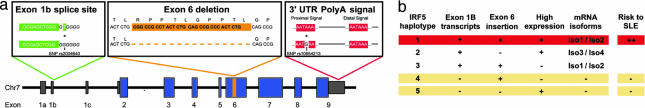
Fig. 2.
Three functional variants in IRF5 define risk and protective haplotypes for SLE. (a) Diagram showing the location of the three common functional alleles identified in IRF5. (b) Summary of IRF5 haplotypes and their association to SLE.
We note that association of the exon 6 indel to SLE was observed only when we conditioned on the exon 1B splice site and polyA+ variants but previously had been masked by the signal of the Group 1 variants in the initial analysis that proceeded in order of statistical significance. Consistent with a model in which the three putative functional alleles (exon 1B, polyA+, and exon 6 indel) are sufficient to explain the observed association to SLE, however, a logistic regression that includes these three variants revealed no additional SNP with P < 0.01. That is, the effect of Group 1 SNPs is statistically indistinguishable from their linkage disequilibrium with the three alleles that have putative functional effects on the structure of IRF5 protein and/or its expression.
Haplotype Analysis Identifies Three Levels of SLE Risk.
To better understand the observed combinations of the three putative functional alleles (and the Group 1 SNPs), we examined the four marker haplotypes defined by: (i) the exon 1B splice site (rs2004640, Group 2), (ii) the polyA+ variant (rs10954213, Group 3), (iii) the exon 6 indel, and (iv) Group 1 (using rs2070197 as a proxy; Table 2). These four variants defined five common haplotypes, each carrying unique combinations of the exon 1B splice site, the exon 6 indel, and the polyA+ variant.
Table 2.
Association of IRF5 haplotypes with SLE
| Haplo-type | Group 2 |
Exon 6 indel | Group 1 |
Group 3 |
T/case frequency* | U/control frequency* | OR (95% c.i.)† | χ2 | P | |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 1B (rs2004640) | rs2070197 | polyA + signal (rs10954213) | ||||||||
| U.S. and United Kingdom | 1 | T | Insertion | C | A | 181 | 99 | 1.90 (1.50–2.41) | 24.2 | 8.5 × 10−7 |
| 2 | T | Deletion | T | A | 248 | 222 | 1.12 (0.93–1.34) | 1.5 | 0.2269 | |
| Trio pedigrees, 555 | 3 | T | Insertion | T | G | 43 | 50 | 0.86 (0.57–1.29) | 0.6 | 0.4384 |
| 4 | G | Insertion | T | G | 195 | 234 | 0.83 (0.69–1.01) | 3.7 | 0.0553 | |
| 5 | G | Deletion | T | A | 104 | 165 | 0.63 (0.50–0.80) | 13.9 | 2.0 × 10−4 | |
| U.S. and United Kingdom | 1 | T | Insertion | C | A | 0.175 | 0.114 | 1.66 (1.40–1.98) | 32.8 | 1.0 × 10−8 |
| 2 | T | Deletion | T | A | 0.377 | 0.363 | 1.06 (0.94–1.21) | 0.9 | 0.3406 | |
| Cases, 1,532 | 3 | T | Insertion | T | G | 0.038 | 0.038 | 1.00 (0.72–1.38) | 0.0 | 0.9981 |
| Controls, 2,878 | 4 | G | Insertion | T | G | 0.290 | 0.351 | 0.76 (0.66–0.87) | 16.4 | 5.3 × 10−5 |
| 5 | G | Deletion | T | A | 0.119 | 0.135 | 0.86 (0.71–1.04) | 2.4 | 0.1233 | |
| Sweden | 1 | T | Insertion | C | A | 0.226 | 0.131 | 1.94 (1.47–2.57) | 21.4 | 3.6 × 10−6 |
| 2 | T | Deletion | T | A | 0.372 | 0.349 | 1.10 (0.89–1.38) | 0.8 | 0.3763 | |
| Cases, 656 | 3 | T | Insertion | T | G | 0.046 | 0.047 | 0.97 (0.59–1.61) | 0.0 | 0.9176 |
| Controls, 718 | 4 | G | Insertion | T | G | 0.219 | 0.296 | 0.67 (0.52–0.85) | 10.4 | 0.0012 |
| 5 | G | Deletion | T | A | 0.137 | 0.177 | 0.73 (0.55–0.99) | 4.3 | 0.0393 | |
| Metaanalysis | 1 | T | Insertion | C | A | 1.78 (1.57–2.02) | 1.4 × 10−19 | |||
| 2 | T | Deletion | T | A | 1.09 (0.99–1.19) | 0.0437 | ||||
| Trio pedigrees, 555 | 3 | T | Insertion | T | G | 0.95 (0.76–1.19) | 0.6743 | |||
| Cases, 2,188 | 4 | G | Insertion | T | G | 0.76 (0.69–0.84) | 5.0 × 10−8 | |||
| Controls, 3,596 | 5 | G | Deletion | T | A | 0.76 (0.67–0.87) | 2.8 × 10−5 |
Bold text refers to the overtransmitted allele; T, exon 1B Splice donor site, T allele shows expression of exon 1B transcripts. Insertion, in-frame insertion/deletion of 30 bp in exon 6 of IRF5, chr7:128,181,324-54 (HG17). Signal variant, “A” allele is associated with short (561-bp) 3′ UTR.
*Number of transmitted (T) and untransmitted haplotypes (U) in pedigrees; frequency of haplotypes in SLE cases and controls.
†OR and 95% confidence intervals (c.i.).
We studied these haplotypes for association to SLE in a large family-based case-control sample totaling 2,188 case and 3,596 control chromosomes (single-marker results, SI Table 11). Haplotype 1 (Table 2) was strongly associated with risk of SLE, appearing on 19.0% of SLE chromosomes in comparison to 11.9% of control chromosomes (P = 1.4 × 10−19; Table 2). In the case-control sample, a single copy of haplotype 1 was associated with an odds ratio (OR) of 1.46, whereas two copies were associated with an OR of 2.96 (SI Table 12). No other IRF5 haplotypes showed positive association with SLE. The high-risk haplotype 1 is predicted to be the only haplotype with the ability to express exon 1B isoforms (because of rs2004640), carries the exon 6 insertion, and is expressed at high levels because of the polyA+ variant.
It has previously been shown that alternative proximal splice acceptors for exon six, termed SS1 and SS2, which are proximate to the exon 6 indel, influence activation of downstream genes (8, 20, 21). As shown in SI Fig. 5, both SS1 and SS2 are used regardless of the exon 6 indel genotype.
Interestingly, although haplotypes 2 and 3 showed no evidence for association to SLE as compared with the overall population (OR = 1.09 and 0.95, P ≥ 0.05, respectively), haplotypes 4 and 5 showed strong evidence for protection. Specifically, each was associated with a ≈25% reduction in risk (OR = 0.76) that was statistically highly significant (P < 5 × 10−8 and 3 × 10−5, respectively). Moreover, individuals that carry Haplotype 1 in trans with either of the haplotypes that lack exon 1B isoform expression (4 and 5) show a reduction in risk of SLE (SI Table 12).
In summary, the highest risk to SLE is observed for a haplotype that is predicted to express at high levels transcripts containing exon 1B and the exon 6 insertion. Haplotypes 2 and 3, which carry only two of the three risk-associated functional alleles, show average risk to SLE. Haplotypes 4 and 5, which carry only one of the three risk-associated functional alleles, and in particular lack exon 1B isoforms, are protective for SLE.
Discussion
Our data provide three contributions regarding the relationship between IRF5 genotype, risk of SLE, and expression of IRF5 isoforms. Specifically, we advance our previous study by showing that (i) in addition to the association of the previously identified exon 1B splice site, at least two independent statistical signals of association to SLE risk can be detected at IRF5, (ii) there are at least two additional functional variants at IRF5, namely the polyA+ signal variant and the 30-bp insertion at exon 6; and (iii) all statistical and functional data can be reconciled in a model in which carrying particular alleles at these three variants (haplotype 1) show strong association to SLE risk, whereas lacking expression of exon 1B isoforms (regardless of the genotype at the other sites) is protective. Whether the three functional alleles of IRF5 interact in an additive or epistatic manner will need to be resolved by in vitro experiments, because all possible allelic combinations of the three functional alleles are not observed in human populations. We also stress that this model is statistically indistinguishable from one in which the Group 1 SNPs are combined with the exon 1B and poly(A)+ SNP and draws any additional support from the functional data derived in vitro and the published role of the exon 6 proline-, glutamic acid-, serine- and threonine-rich domain on function in IRF family members (14).
These alterations in IRF5 protein structure and expression levels presumably shape the IRF5 transcriptional cascade and downstream immune responses. It has been shown that IRF5 is activated by Toll-like receptors 7/9 and type 1 IFN signaling and that, upon stimulation, different IRF5 protein isoforms differentially activate transcription of target proinflammatory cytokines (8, 20, 21). We therefore propose that the differential expression of IRF5 target genes conferred by IRF5 genotype may modify the immune response in a manner that predisposes to SLE, perhaps in response to chromatin-containing Ig immune complexes and type-I IFN, both of which are elevated in the blood of SLE patients and may stimulate IRF5. Identifying the specific nature of the altered IRF5 transcriptional response and finding interventions that shift it toward that caused by the low-risk exon 1B lacking isoforms (haplotypes 4 and 5) seems a well-motivated approach to prevention and treatment of SLE.
Finally, these results have general implications for the genetic analysis of complex traits. Candidate-gene and genome-wide analyses may identify a particular SNP as reproducibly associated to disease, highlighting a region likely to harbor variants associated with disease pathophysiology. By design, the initial screens are incomplete assessments of variation present at a locus. A full characterization of genetic variation at associated loci in large samples is required to refine models of genotype-phenotype correlation. Our results make clear that (i) there may be multiple functional variants, (ii) the most strongly associated SNP may be not the causal variant but rather a proxy for a haplotype of multiple variants, and (iii) related phenotypes such as gene expression may help resolve the functional role of alleles. More generally, there is much to learn about the genetic architecture of complex traits that will inform the search for causes of disease.
Materials and Methods
Expression of IRF5 in whole blood and cell lines was measured by qPCR and Northern and Western blotting, as described in SI Text. IRF5 message stability was measured by using transient transfection and RNA decay assays, described in detail in SI Text. Normalized IRF5 mRNA expression levels were obtained from data made available by the GENEVAR project at the Sanger Centre from EBV-transformed B cells derived from the 270 HapMap samples (IRF5 exon 9 probe GI_38683858-A). In addition, IRF5 expression values (probeset 205469_s_at) were obtained from a data set of 233 CEPH EBV-transformed B cell lines (16, 17) (GEO accession no. GSE1485). Association of genotype to IRF5 expression levels and conditional logistic regression analyses were conducted using WHAP (http://pngu.mgh.harvard.edu/purcell//whap).
A collection of samples of European descent were genotyped by using the primers and probes described in SI Table 13. The family samples used were collected at the University of Minnesota and Imperial College (22–25). In addition, independent population-based samples from the University of Minnesota, Imperial College, and the University of California, San Francisco, Lupus Genetics Project collection (26) and 1,439 controls from the New York Cancer Project (27) were tested. The study also included 338 SLE patients from Sweden, 213 of them recruited at the Karolinska Hospital in Stockholm (28) and 125 at Uppsala University Hospital (6), and 363 healthy age- and sex-matched controls from the same geographical regions as the SLE patients. The SLE patients fulfilled the American College of Rheumatology revised criteria for SLE (29). In addition, 270 samples from the International Haplotype Map project (15) and 233 CEPH individuals (14 extended pedigrees, including 21 trios that are part of the HapMap CEU samples, and 38 unrelated individuals) described by Morley et al. (17) were genotyped for IRF5 region markers, using primers and methodology described in SI Text. The Human Genome Diversity Panel was genotyped to assess the frequency of IRF5 alleles in world populations as described in SI Fig. 6 and SI Table 14. IRF5 was resequenced in eight controls and 40 SLE cases collected at Uppsala, Sweden, using 23 PCR fragments that covered 1 kb upstream of exon 1A and all exons and introns. In addition, all exons of IRF5 1 kb upstream of exon 1A were resequenced in 96 SLE cases of European descent from the Minnesota SLE cohort, as described in SI Text. Family-based and case/control association analyses, including permutation testing, were conducted by using Haploview v3.3 (30). Conditional logistic regression analyses of single markers and haplotypes were performed by using the WHAP software program. Haplotypic association results in the family-based U.S. and United Kingdom cohort, the case-control cohort collected in the U.S. and United Kingdom, and the Swedish case-control cohort were combined by using the Mantel–Haenszel metaanalysis of the ORs (31, 32).
Supplementary Material
Acknowledgments
We thank Ann-Christine Wiman and Lili Milani for excellent technical assistance with genotyping in the Swedish cohort. This work was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (T.W.B. and P.R.B.); the National Institute of Arthritis, Musculoskeletal and Skin Diseases, NIH (R.R.G., E.C.B., D.A., T.W.B., and L.A.C.); the Alliance for Lupus Research (T.W.B.); the Lupus Research Institute (T.W.B.); the Mary Kirkland Center for Lupus Research (T.W.B. and L.A.C.); the Lupus Foundation of Minnesota (T.W.B.); the Swedish Research Council for Medicine (A.-C.S and L.R); the Knut and Alice Wallenberg Foundation (A.-C.S); the Swedish Rheumatism Foundation (L.R.); the King Gustaf V 80 Years Foundation (L.R. I.G., and E.S.); the Center of Gender Medicine Karolinska Institutet (E.S.); the Swedish Heart-Lung Foundation (E.S); and the Lymphoma Research Foundation (I.A.V.). The Hopkins Lupus Cohort is supported by NIH Grant AR 43727, and the Johns Hopkins General Clinical Research Center is supported by NIH Grant M01-RR-00052. These studies were also supported in part by the General Clinical Research Center, Moffitt Hospital, University of California, San Francisco, with funds provided by the National Center for Research Resources, Grant 5 M01 RR-00079, U.S. Public Health Service.
Abbreviations
- SLE
systemic lupus erythematosus
- indel
insertion/deletion
- CEPH
Centre d'Etude du Polymorphisme Humain
- CEU
CEPH (Utah residents with ancestry from northern and western Europe)
- IRF
IFN regulatory factor
- OR
odds ratio.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701266104/DC1.
References
- 1.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, et al. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 3.Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 4.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, et al. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 6.Sigurdsson S, Nordmark G, Goring HH, Lindroos K, Wiman AC, Sturfelt G, Jonsen A, Rantapaa-Dahlqvist S, Moller B, Kere J, et al. Am J Hum Genet. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes BJ, Moore PA, Pitha PM. J Biol Chem. 2001;276:23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 8.Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. J Biol Chem. 2004;279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- 9.Honda K, Yanai H, Takaoka A, Taniguchi T. Int Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 10.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 11.Rhodes DA, Trowsdale J. Rev Immunogenet. 1999;1:21–31. [PubMed] [Google Scholar]
- 12.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. J Am Med Assoc. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 13.Klos KL, Sing CF, Boerwinkle E, Hamon SC, Rea TJ, Clark A, Fornage M, Hixson JE. Arterioscler Thromb Vasc Biol. 2006;26:1828–1836. doi: 10.1161/01.ATV.0000231523.19199.45. [DOI] [PubMed] [Google Scholar]
- 14.Levi BZ, Hashmueli S, Gleit-Kielmanowicz M, Azriel A, Meraro D. J Interferon Cytokine Res. 2002;22:153–160. doi: 10.1089/107999002753452764. [DOI] [PubMed] [Google Scholar]
- 15.International HapMap Consortium. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conne B, Stutz A, Vassalli JD. Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 19.Edmonds M. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 20.Mancl ME, Hu G, Sangster-Guity N, Olshalsky SL, Hoops K, Fitzgerald-Bocarsly P, Pitha PM, Pinder K, Barnes BJ. J Biol Chem. 2005;280:21078–21090. doi: 10.1074/jbc.M500543200. [DOI] [PubMed] [Google Scholar]
- 21.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Mol Cell Biol. 2002;22:5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaffney PM, Kearns GM, Shark KB, Ortmann WA, Selby SA, Malmgren ML, Rohlf KE, Ockenden TC, Messner RP, King RA, et al. Proc Natl Acad Sci USA. 1998;95:14875–14879. doi: 10.1073/pnas.95.25.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaffney PM, Ortmann WA, Selby SA, Shark KB, Ockenden TC, Rohlf KE, Walgrave NL, Boyum WP, Malmgren ML, Miller ME, et al. Am J Hum Genet. 2000;66:547–556. doi: 10.1086/302767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham DS, Wong AK, McHugh NJ, Whittaker JC, Vyse TJ. Hum Mol Genet. 2006;15:3195–3205. doi: 10.1093/hmg/ddl395. [DOI] [PubMed] [Google Scholar]
- 25.Graham RR, Langefeld CD, Gaffney PM, Ortmann WA, Selby SA, Baechler EC, Shark KB, Ockenden TC, Rohlf KE, Moser KL, et al. Arthritis Res. 2001;3:299–305. doi: 10.1186/ar319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsa A, Peden E, Lum RF, Seligman VA, Olson JL, Li H, Seldin MF, Criswell LA. Genes Immun. 2002;1(3) Suppl:S42–S46. doi: 10.1038/sj.gene.6363907. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. J Urban Health. 2004;81:301–310. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svenungsson E, Gunnarsson I, Fei GZ, Lundberg IE, Klareskog L, Frostegard J. Arthritis Rheum. 2003;48:2533–2540. doi: 10.1002/art.11264. [DOI] [PubMed] [Google Scholar]
- 29.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 32.Woolson RF, Bean JA. Stat Med. 1982;1:37–39. doi: 10.1002/sim.4780010106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.