Abstract
Vocal learning and neuronal replacement have been studied extensively in songbirds, but until recently, few molecular and genomic tools for songbird research existed. Here we describe new molecular/genomic resources developed in our laboratory. We made cDNA libraries from zebra finch (Taeniopygia guttata) brains at different developmental stages. A total of 11,000 cDNA clones from these libraries, representing 5,866 unique gene transcripts, were randomly picked and sequenced from the 3′ ends. A web-based database was established for clone tracking, sequence analysis, and functional annotations. Our cDNA libraries were not normalized. Sequencing ESTs without normalization produced many developmental stage-specific sequences, yielding insights into patterns of gene expression at different stages of brain development. In particular, the cDNA library made from brains at posthatching day 30–50, corresponding to the period of rapid song system development and song learning, has the most diverse and richest set of genes expressed. We also identified five microRNAs whose sequences are highly conserved between zebra finch and other species. We printed cDNA microarrays and profiled gene expression in the high vocal center of both adult male zebra finches and canaries (Serinus canaria). Genes differentially expressed in the high vocal center were identified from the microarray hybridization results. Selected genes were validated by in situ hybridization. Networks among the regulated genes were also identified. These resources provide songbird biologists with tools for genome annotation, comparative genomics, and microarray gene expression analysis.
Keywords: cDNA microarray, developmental gene expression, EST sequencing, microRNA, zebra finch
The genomes of many species, including human, mouse, chicken, Drosophila, Caenorhabditis elegans, and zebrafish, have been sequenced, with the result that genomic, computational, and microarray tools have greatly accelerated discoveries in many fields of biology. However, the availability of such resources has lagged in songbirds, even as two species, the zebra finch (Taeniopygia guttata) and the canary (Serinus canaria), have received much attention from scientists studying the biology of vocal learning and neuronal replacement in the adult brain. Songbirds have been proven to be exceedingly favorable material for studying the basic biology of these two phenomena. Here we describe our effort to develop molecular resources that can be used in such work, including the large-scale sequencing of ESTs and the printing of cDNA microarrays of genes expressed in zebra finch brains. The high vocal center (HVC) controls song learning in juveniles and song production in adult birds and is also a place where neuronal replacement occurs in adulthood (1). We used microarrays to profile genes differentially expressed in the HVC of adult male zebra finches and canaries, compared with universal total brain RNA references, and validated the microarray results by in situ hybridization.
Results and Discussion
Libraries, Clones, and Sequence Analysis.
We made four cDNA libraries from zebra finch brains at successive developmental stages, mindful that the most prominent structural and functional brain changes occur during development. The embryonic cDNA library used brains collected from embryos of both male and female birds at embryonic days 5–13 (hatching is on day 14). The three developmental libraries were generated by grouping total brain tissues of male zebra finches at posthatching days 1, 3, 5, 10, 20, 30, 40, 50, 60, 70, and 90 (Table 1). These four libraries cover the entire period of brain development up to sexual maturity (at approximately day 90), including the period during which juvenile zebra finches learn their song. Standard library construction procedures were used to make the cDNA libraries. In total, 11,000 cDNA clones from these libraries were sequenced (one read) from the 3′ ends, yielding 700- to 800-nt readable sequences. After eliminating contaminated clones, sequences shorter than 100 nt, and failed sequences, a total of 9,845 successful sequence reads were obtained.
Table 1.
Developmental stages for library construction
| Libraries | Developmental stages, days |
|---|---|
| Embryonic | Embryonic 5, 6, 7, 8, 9, 10, 11, 12, and 13 |
| Developmental I | Posthatching 1, 3, 5, 10, and 20 |
| Developmental II | 30, 40, and 50 |
| Developmental III | 60, 70, and 90 |
The table indicates the developmental stages at which brains were collected for cDNA library construction. For the embryonic brain library, both male and female brains (total N = 30) were used. For all other libraries, one brain was obtained for each of the specific ages indicated above by using only male brains.
The 9,845 sequence reads represent 5,866 distinct gene transcripts (Table 2). Annotations of the clones are based on sequence homology with genes identified in other species. We compared our clone collection with sequences available in several public databases. Blast searches against the National Center for Biotechnology Information (NCBI) databases resulted in 2,802 (47.7%) clones with matching cDNA sequences and 1,741 (29.7%) clones with matching protein sequences. Blastn search against the The Institute for Genomic Research (TIGR) chicken cDNA database resulted in 3,294 (56.1%) clones with matching nucleotide sequences. Blastx search against the SWISS-PROT protein database resulted in 1,374 (23.4%) clones with matching protein sequences. Blastn search against the NCBI annotated chicken genome sequences resulted in 1,893 (32.2%) hits matching the annotated gene sequences. In all, 65% of the distinct zebra finch sequences matched annotated sequences in the databases. Many (35%), however, did not, possibly because their 3′ end sequences fall in the 3′ end untranslated regions of genes, which are less conserved across species than protein-coding regions. Consequently, many transcripts may have failed to match orthologs in other species. Nonetheless, some of the new sequences may represent previously unrecognized gene transcripts.
Table 2.
Gene diversity across developmental stages
| Libraries | No. of sequences | No. of distinct genes | Gene discovery rate, % | No. of singletons | Percentage of singletons |
|---|---|---|---|---|---|
| Embryonic | 2,920 | 1,929 | 66.1 | 1,056 | 36.2 |
| Developmental I | 3,566 | 2,593 | 72.7 | 1,581 | 44.3 |
| Developmental II | 1,873 | 1,611 | 86.0 | 989 | 52.8 |
| Developmental III | 1,486 | 1,158 | 77.9 | 555 | 37.4 |
| Total | 9,845 | 5,866 | 59.6 | 4,181 | 42.5 |
Within each library, distinct genes refer to clones that represent different sequences. Gene discovery rate = (number of distinct genes)/(number of sequenced clones in a library). Singletons refer to the clones that appear as a single copy and do not match any other clones in any of the libraries. Percentage of singletons = (number of singletons)/(number of sequenced clones in that library). The gene discovery rate is significantly higher in Developmental Library II than in each of the other libraries (Fisher's exact test; P < 10−8 in all cases). The comparison between Developmental Libraries II and III, which contain similar numbers of sequences, is significant, P < 1.2 × 10−9.
Recently, two other songbird EST sequencing efforts have been carried out at Duke University (http://songbirdtranscriptome.net) (2) and the University of Illinois (http://titan.biotec.uiuc.edu/songbird) (3). To identify the overlap of our sequences with these two data sets, we first compared our 5,866 distinct zebra finch ESTs to the ≈4,750 distinct 3′ sequences generated by the Duke University group. Using the same Blastn criteria that we used to identify redundant clones (e value ≤ 1e−50, and ≥90% nucleotide identity), we found that ≈1,950 of our sequences matched sequences in the Duke data set, suggesting that they may be transcripts of the same genes. The remaining 3,912 distinct sequences in our dataset were not identified by the Duke group. Because the ≈36,000 ESTs from the University of Illinois were sequenced from 5′ ends, we were not able to do the overlap comparison with this data set without genomic sequence information.
Redundancy Analysis of Clones from Nonnormalized Libraries.
We performed redundancy analysis by comparing clones pairwise, on the basis of the 700- to 800-nt sequences at the 3′ ends. The 9,845 sequenced clones represent a total of 5,866 different gene transcripts, a gene discovery rate of 59.6%. The actual number of distinct genes may be slightly smaller, because splicing variants from the same gene but with different 3′ terminal sequences would have been counted as different gene transcripts. A large number of the sequenced clones (4,181, 42.5%) are singletons, and only a handful of highly redundant genes are represented between 50 and 100 times (Table 3). Normalization procedures have often been used in EST sequencing projects to remove highly redundant gene copies and thus increase the gene discovery rate (4). We did not normalize the cDNA libraries for two reasons. First, normalization procedures require extensive manipulation of cDNA, resulting in potential DNA damage and loss of low copy number genes. Second, hybridization-based normalization procedures could lead to the loss of closely related but different gene sequences. Our results show that the nonnormalized libraries present a very high gene discovery rate. The high gene discovery rate and the high percentage of singletons indicate the high quality of our cDNA libraries and suggest that random clone sequencing can yield a large number of distinct genes.
Table 3.
Redundancy analysis
| Frequency of clones | No. of clones | No. of genes |
|---|---|---|
| 1 | 4,181 | 4,181 |
| 2 | 1,918 | 959 |
| 3 | 929 | 310 |
| 4 | 608 | 152 |
| 5 | 326 | 65 |
| 6 | 195 | 30 |
| 7 | 161 | 23 |
| 8 | 180 | 23 |
| 9 | 153 | 17 |
| ≥10 | 1,195 | 106 |
| Total | 9,845 | 5,866 |
Redundancy analysis was performed by blasting every clone against every other clone. Sequences of all clones from all cDNA libraries were combined for this analysis.
Gene discovery rates for the four cDNA libraries are shown in Table 2. Among the four libraries, the developmental II library (30- to 50-day-old brains) shows the highest gene discovery rate and the highest number of singletons. The 1,873 clones sequenced from this library represent 1,611 distinct gene transcripts (86.0% gene discovery rate) and 989 (52.8%) singletons. In contrast, the 2,920 clones obtained from the embryonic library represent 1,929 distinct genes (66.1% gene discovery rate) and 1,056 (36.2%) singletons. Although the gene discovery rate and the percentage of singletons would be expected to decrease as more clones are sequenced, the difference in gene discovery rates among the libraries may also reflect the diversity of gene expression during developmental stages. The developmental II library, with the highest gene discovery rate, covers the time when juvenile zebra finches become independent and song learning starts (5). Marked changes in the song system as well as in other brain regions during this time may require diverse gene expression programs.
Distribution of Functional Gene Groups Among Developmental Stages.
Sequencing of ESTs can be used for gene expression profiling. Because our cDNA libraries were not normalized, gene representation among the libraries reflects relative gene expression levels during different developmental stages. We grouped sequences on the basis of their functional annotations as per the Gene Ontology (GO) database (6). Many functional gene groups show distinct developmental regulation patterns (Fig. 1). For example, gene groups related to protein synthesis, transcription, RNA processing, and cell cycle are highly expressed during the embryonic stage, suggesting that macromolecule synthesis dominates during this stage of brain development. Twice as many genes related to RNA splicing and processing are found during this stage compared with later developmental stages, suggesting that posttranscriptional regulation of gene expression plays a prominent role in embryonic brain development.
Fig. 1.
Developmental representation of functional groups of genes. Genes were classified by associating GO terms to clones with UniGene IDs. The analysis shown here used only clones for which UniGene IDs were available. The figure shows the number of clones associated with GO terms per 1,000 in each library for a selected subset of functional groups. In each functional group, the starred column indicates the library with the highest clone count. In each case, this count was significantly higher than the average of all four libraries (Fisher's exact test; P < 0.05 in each case).
During postembryonic brain development, neurons differentiate and form connections, and neural circuits are fine-tuned by experience and learning. During this time, apparently, the profile of gene expression changes, too. Our results suggest that genes related to synaptic transmission and neuron development become more highly expressed in the posthatching libraries (Fig. 1). Moreover, apoptosis-related genes are found at higher levels during posthatching days 1–20 compared with other developmental stages. Because the cDNA libraries were made from entire brains, the higher expression of apoptosis genes could relate to intense cellular winnowing in a few specific brain regions or to more widespread programmed cell death. In the adult songbird brain, new neurons continue to be produced and recruited into many brain regions, and many of the new cells die within the first few weeks after birth (7–9). The extent to which neuronal apoptosis occurs during development and adulthood could be an interesting area for future research.
Identification of Zebra Finch MicroRNAs.
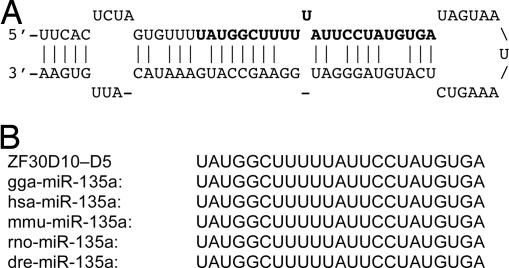
MicroRNAs are a class of ≈21-nt-long non-protein-coding RNA molecules that regulate gene expression posttranscriptionally. Some microRNAs also play essential roles in the regulation of neuronal differentiation and neuronal cell fate determination (10, 11). We compared our ESTs with all currently known animal microRNA sequences and identified seven ESTs that display high sequence homology with five microRNAs: miR-56, miR-87, miR-135a, miR-297, and miR-466. All of these ESTs contain a region that can form a hairpin-shaped secondary structure, a typical feature of microRNA precursors; the conserved microRNA sequences are found within the stem regions of the hairpin structures. As shown in Fig. 2, one microRNA, miR-135a, has identical sequences in chicken, human, mouse, rat, zebrafish, and zebra finch, suggesting that this microRNA might have highly conserved functions [sequences of other putative miRNAs can be found in supporting information (SI) Fig. 5]. The expression of miRNAs during different developmental stages suggests that miRNAs may play regulatory roles in songbird brain development. miR-135a is expressed in human and mouse brains, but not in other tissues (12, 13). It has recently been shown that miR-135a expression is induced when embryonic carcinoma cells treated with retinoic acid differentiate into neurons (13), suggesting that it has a role in neural differentiation and neuronal fate determination.
Fig. 2.
The predicted hairpin-like secondary structure of clone zf30d10-D5 (EE053401) and its sequence homology with miR-135a known from other species. (A) The predicted precursor hairpin structure within clone zf30d10-D5. The 23-nt miRNA sequence is highlighted in bold. Vertical bars indicate complementary nucleotides. (B) Sequence alignment of the predicted miRNA sequence of zf30d10-D5 with miR-135a from other species. gga, Gallus gallus; hsa, Homo sapiens; mmu, Mus musculus; rno, Rattus norvegicus; dre, Danio rerio.
Target prediction analysis resulted in ≈290 potential target genes for the five miRNAs (a list of the predicted target genes is provided in SI Table 4), on the basis of analysis of the 3′-untranslated regions of annotated chicken genes from the NCBI databases. Among the potential target genes, 27 are found in our EST collection, and 9 are coexpressed in the same developmental libraries as their corresponding miRNAs. Transcripts of some transcription factors and neuronal specific genes, such as neuropilin, neuroepithelial cell transforming gene 1, and neurotensin precursor, are among the putative miRNA targets. Our observations are compatible with the possibility that miRNAs and their target genes play important roles in songbird brain development.
Microarray Analysis of Genes Differentially Expressed in HVC.
The forebrain nucleus HVC controls song production in songbirds (14, 15). One of its neuron types undergoes replacement in adulthood (16, 17). We printed zebra finch cDNA microarrays, and profiled gene expression in the HVC of adult male zebra finches. A pan-finch probe, made of adult male zebra finch whole-brain RNA, was used as a reference. The software package SAM (Statistical Analysis of Microarrays) was used for data analysis (18). At FDR = 0 (0% false discovery rate), 17 genes were up-regulated and 17 genes were down-regulated in HVC, compared with the reference. At FDR ≤ 5%, 404 genes were up-regulated and 408 were down-regulated. The up-regulated genes include parvalbumin (EE056773), calmodulin binding transcription factor (EE049904), histidine triad protein Hint1 (EE050138), calcineurin (EE058025), and ubiquitin carboxyl-terminal hydrolase L1 (EE050470). The down-regulated genes include Purkinje cell protein 4 (PCP-4) (a regulator of calmodulin; EE049987), cAMP-regulated phosphoprotein (EE049891), kainate receptor (EE056631), reelin (EE054247), and MHC class I protein (EE060264). A list of differentially expressed genes at FDR ≤ 5% is shown in SI Table 5. Notably, several of the differentially regulated genes (both up- and down-regulated), such as parvalbumin, calmodulin-binding transcription factor (CaM-TF), calcineurin, and PCP-4, are related to calcium signaling pathways. Calcium plays important roles in the functions of the nervous system, especially in synaptic transmission and neuronal electrical activity. In particular, the neuronal firing patterns in HVC, which controls song behavior, are thought to be intimately related to intracellular calcium (19). Because calcium plays such a basic role in neuronal excitation, it is hard to imagine how HVC neurons would be different from those in the rest of the brain, yet the observation of differential expression in HVC of calcium-related genes is robust.
Validation of the Microarray Results by in Situ Hybridization.
We used in situ hybridization to validate the microarray hybridization results for a handful of differentially expressed candidate genes. Fig. 3 shows the prominent expression in HVC of four up-regulated genes, parvalbumin (EE056773), CaM-TF (EE049904), RGS4 (EE055184), and cornified envelope protein (EE055259) and the low or absent expression of Reelin (EE054247) and PCP-4 (EE049987). In the microarray experiment, differentially expressed genes were identified by comparing expression in HVC with total-brain RNA, so it is not surprising that these genes were also expressed in brain areas other than HVC. In addition, in situ hybridization revealed that some of the candidate genes were also expressed in other song nuclei. For example, parvalbumin showed relatively high expression in HVC, robust nucleus of the archistriatum (RA), and lateral magnocellular nucleus of the anterior neostriatum (lMAN), whereas Reelin was down-regulated in these three nuclei. The song system nuclei, which are functionally related (20), may share similar expression patterns for some but not all genes.
Fig. 3.
In situ hybridization performed on zebra finch and canary brain sections to validate a selected set of differentially expressed genes identified by microarray hybridization. (A) Genes expressed at higher levels in the HVC and, in some cases, also in other song nuclei of adult male zebra finches. (B) Genes expressed at higher levels in the HVC of adult male canaries. (C) Genes down-regulated in the HVC of adult male zebra finches (left two sections) and canaries (right two sections). Parvalbumin (EE056773); Cal-TF, Calmodulin-binding transcription factor (EE049904); RGS4, regulator of G protein signaling 4 (EE055184); Corn Eve, cornified envelope protein (EE055259); Reelin (EE054247); PCP-4, Purkinje cell protein 4 (EE049987).
Parvalbumin, a calcium-binding protein, is commonly expressed in inhibitory interneurons. Parvalbumin protein has been detected in interneurons in HVC by immunological studies and is thought to play a role in establishing the inhibitory microcircuits in the song system during juvenile song learning (19). A microarray gene profiling experiment has recently shown that parvalbumin is down-regulated in the visual cortex of dark-reared animals (21); it is thought that maturation of the parvalbuminergic inhibitory circuit is associated with the critical period of visual cortex development (22). It would be of interest to know how parvalbumin is regulated during song circuit development, what determines when and in which neurons it is expressed, and how its expression responds to sensory/motor learning and experience. We remain very intrigued, too, by the selective expression of the cornified envelope protein in HVC. It has been reported that cornified envelope protein is involved in epidermal differentiation, and that its mutation causes skin diseases (23). Its functional role in the central nervous system has yet to be explored.
Comparisons with Another Songbird Species.
The canary is another oscine songbird that has been extensively used for laboratory studies of vocal learning (24) and neuronal replacement (25). To explore the applicability of our zebra finch cDNA microarrays to the canary, we profiled gene expression in the HVC of adult male canaries. A pan-canary probe made of whole-brain RNA of adult male canaries was used as a reference. Different but overlapping sets of genes were regulated in the HVC of canaries and zebra finches. For example, at FDR ≤ 5%, 193 of the 404 genes up-regulated in the finch HVC were also up-regulated in the canary HVC, and 190 of the 408 genes down-regulated in the finch HVC were also down-regulated in the canary HVC. A complete list of differentially expressed genes (FDR ≤ 5%) in canaries can be found in SI Table 6. Differences in gene expression in the HVC of adult male canaries and zebra finches could have several explanations. First, gene expression patterns in the HVC of the two species might be intrinsically different. Second, canaries, unlike zebra finches, are seasonal birds; their breeding behavior and physiology change seasonally. The male canaries used in the experiment were collected during the spring breeding season. During this time, it is known that, in addition to other physiological changes, the blood testosterone level is high, which may influence gene expression in HVC and in other parts of the brain. Nonetheless, despite these factors, many genes were differentially regulated in the same direction in both species.
In situ hybridization was performed to validate the differentially expressed genes in the canary, with the same set of probes that were used for the zebra finch. Parvalbumin, CaM-TF, RGS-4, and cornified envelope protein were up-regulated in the HVC of both finch and canary, whereas Reelin and PCP-4 were down-regulated in the two species (Fig. 3). Not only were these genes regulated similarly in the canary and zebra finch HVC, but they also showed similar expression patterns in other song system nuclei and/or other brain regions of the two species. Taken together, our results suggest that our zebra finch cDNA microarray can be used effectively to study gene expression in canaries and maybe in other songbird species.
Bioinformatic Analysis of Differentially Expressed Genes.
We used the bioinformatics tool Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA) to explore how individual differentially expressed genes interrelate or interact with each other. Of the 812 genes differentially expressed in the zebra finch HVC, 147 are present in the IPA database. Many of these genes are members of known gene networks related to various cell functions. The most significant network revealed by our analysis, shown in Fig. 4, has an IPA score of 24 and contains 18 genes predicted to be part of an interacting gene network. Of these, 4 are related to protein translation and 10 to transcription and mRNA processing. These two clusters would be expected from the IPA analysis to function in an interconnected manner, but this prediction is based on published material from other systems and would have to be confirmed experimentally in the zebra finch. The names of the 18 genes in this putative network can be found in SI Table 7. The protein translation machinery components (EIF3S2, 5, 6, and EIF5) may be under transcriptional and posttranscriptional regulation by the transcription and mRNA processing factors. Conversely, the protein synthesis of the transcriptional regulators may be under the control of the translational proteins (Fig. 4). Interestingly, one member and also a possible target of this network is Contactin 4 (CNTN4), an axon-associated cell adhesion molecule that plays a central role in neuronal differentiation, axonal growth, and circuit formation (26, 27). The 3p-deletion syndrome, a genetic defect associated with both verbal and nonverbal developmental delays in humans, has been correlated to chromosomal disruption of CNTN4 (28).
Fig. 4.
The most significant network of genes differentially expressed in the zebra finch HVC includes two biological themes, gene expression (right-hand cluster) and protein translation (left-hand cluster), as identified by IPA. Each gene is represented by a symbol and its abbreviation; only genes with shaded symbols were, in our data, differentially expressed in HVC. The name of each gene and its known function appear in SI Table 7. Gene groups represented by the symbols are indicated on the lower right. Solid lines linking genes indicate a direct action, and broken lines indicate an indirect action. Lines with arrows indicate that one gene acts on the other, and lines without arrows indicate that the corresponding proteins interact with each other. The blank symbols pertain to genes that were either not present in our array or not differentially expressed. This figure shows the complexity of the interrelations uncovered by combining microarray data and bioinformatics analyses.
Conclusions
Our work has yielded the following valuable resources for scientists working on the neurobiology, molecular biology and behavior of songbirds: (i) EST sequences, grouped by developmental stage and functionally annotated; (ii) cDNA clones; and (iii) cDNA microarrays representing 5,866 unique gene transcripts. In addition, our sequencing and functional annotation of 3′ end sequences of cDNA clones from nonnormalized developmental libraries have yielded a rich set of sequences whose expression seems to differ between developmental stages, providing potential insights into patterns of gene expression at different stages of songbird brain development. The 30- to 50-day library shows the greatest diversity of gene expression. This may relate to the marked changes in lifestyle and behavior of juveniles during this time. Song development starts soon after day 30; at the same time, juveniles become independent of their parents and must forage for their own food. Moreover, as the family group dissolves, the juveniles form new associations. We do not know to what extent these changes affect brain circuitry, although we do know that the brain pathways involved in song learning are growing and connecting during the very time that song learning takes place (29–31). We have also identified five miRNAs among the 3′ EST sequences expressed during songbird brain development. We have used cDNA microarrays to profile gene expression levels in the song nucleus HVC of adult male zebra finches and have validated the results by in situ hybridization. We have also verified the usefulness of our zebra finch microarray for gene expression analysis in canaries. At the procedural level, we have established that total brain RNA can be a convenient universal reference for cDNA microarray experiments, so that results from different microarray experiments done at different times or in different laboratories can be cross-compared. Our work complements other songbird EST sequencing and microarray initiatives, and greatly extends the quantity of genes that are now available for future study. As in other work that has produced genomic resources for a particular research community, the tools we offer will allow many hypotheses (e.g., about molecular mechanisms underlying song learning and neuronal replacement) to be tested in the songbird brain.
A Multipurpose Automated Genome Project Investigation Environment (MAGPIE) database (32), containing all sequence and annotation data for the 9,845 ESTs, can be accessed at (http://magpie.ucsd.edu/magpie/zfinch_v1). It is equipped with functions such as keyword search, clone name search, and blast search. Sequence data have been deposited in the NCBI database under accession numbers EE049699–EE062017. Individual cDNA clones and microarrays are available to the research community upon request.
Methods
Animals, Library Construction, and Sequencing.
Birds were reared in our aviary under a typical 12-h light/12-h dark cycle. All birds behaved normally. Brains were collected sometime during the morning, after the birds woke up. Birds killed before day 31 were housed in the same cage with parents and siblings; those killed after that time were housed in a communal cage with other males, both juveniles and adults. Entire brains were obtained from zebra finches at various developmental stages. Messenger RNA was purified. An oligo(dT) primer [5′-atat-GCGGCCGCNotI-AG-T18-(A, C, G,)-3′] was used to make standard cDNA libraries (SuperScript Choice System; Invitrogen, Carlsbad, CA), and the cDNA inserts were ligated to the EcoRI and NotI sites of the plasmid vector pBluescript. The quality of the libraries was assessed by restriction digestion, PCR, and gel electrophoresis. Typically, >95% of the clones contained cDNA inserts, with an average insert size of 1.5–2.5 kb. Clones from the libraries were randomly picked, and plasmid DNA was purified and sequenced from the 3′ ends one read by using the T3 primer, yielding ≈700-nt readable sequences.
Sequence Analysis.
Initial sequence analysis and construction of a web-based database were performed by using the MAGPIE software package (32). Vector sequences and unsuccessful sequencing products were removed. The remaining ESTs were compared pairwise to identify redundant clones. Any two sequences showing ≥90% homology with an expectation value ≤1e−50 were considered redundant; otherwise, they were considered as distinct gene transcripts. The nonredundant ESTs were then compared, by using the Blastn and Blastp programs, with the NCBI nucleotide and protein databases, with the TIGR chicken gene index, and with the NCBI's human, mouse, and chicken UniGene databases. Nucleotide sequences matching with e values ≤ 1e−20 and protein sequences matching with e values ≤ 1e−10 were considered to be homologous with the query sequences.
MicroRNA Identification and Target Prediction.
Zebra finch microRNAs were identified by comparing all EST sequences with known microRNA sequences collected in Rfam (http://microrna.sanger.ac.uk/sequences/index.shtml). EST sequences with high homology (>16 identical nucleotides) to known microRNAs were selected, and the adjacent regions were extracted and examined for secondary structure by using the software package MFOLD (33). Sequences with high homology to known microRNAs and with a hairpin-like precursor structure were considered zebra finch microRNAs (34). Putative microRNA target genes were predicted by searching for miRNA complementary sequences within 3′ UTRs of chicken mRNAs by using a modified Smith–Waterman nucleotide alignment program (35). Chicken mRNAs whose 3′ UTRs had at least one region complementary to the first eight nucleotides of the 5′ end of the miRNA, with at most one insertion or two mismatches, were selected as candidate miRNA target genes.
Microarray Printing.
On the basis of the redundancy analysis, highly redundant clones were removed, so that all gene transcripts were represented one or two times in the set used for micorarray printing. The cDNA inserts were PCR amplified. The PCR products were purified, checked by gel electrophoresis and quantified by UV absorption. The DNA samples were dissolved in 35% DMSO and printed in duplicate onto glass slides (UltraGAPS; Corning, Midland, MI) using the MicroGrid (BioRobotics, Cambridge, U.K.). Slides were then cross-linked by UV radiation at 360 J.
Microarray Hybridization.
Zebra finch and canary frozen brains were cut at 80-μm intervals, and the HVC areas were dissected under a dissection microscope. Total RNA was purified (RNAeasy; Qiagen, Valencia, CA) and amplified one round (36). Total RNA from entire adult male zebra finch or adult male canary brains was also amplified one round and used as a reference probe. Amplified RNA (2 μg) was used to make cDNA probes with Cy3-dUTP or Cy5-dUTP. Reverse transcription was carried out at 42°C for 2 h, followed by RNase H digestion (2 units) for 30 min at 37°C. The labeled probes were purified, combined, and hybridized to the array. Three birds were used for each group, processed individually, and two hybridizations were performed for each bird with dye reversal.
Data Processing and Analysis.
Hybridized arrays were scanned at 647 nm and 555 nm (ScanArrayGx; PerkinElmer, Boston, MA). Spot images were quantified by using the software packages GenePix and GeneTraffic. The spot intensity data were Lowess-normalized. Because of duplications in printing and in hybridization, each gene was represented with at least four spots per bird. A spot was counted as valid provided its signal intensity was at least twice the background. The data for a given gene were accepted for further analysis if at least two of its four spots were valid in at least two of the three birds. The log2 ratios of the experimental to reference pixel intensities were averaged over the valid spots for each bird. A one-class t test was performed by using Significance Analysis of Microarrays (SAM), with default settings of random seed number and 100 permutations for FDR analysis.
The genes with FDR ≤ 5% were considered significantly differentially expressed and subjected to further analysis using IPA on the basis of associated annotations in the GO database (12). Because the GO database currently does not include zebra finch clones, we blasted our clones by using Blastn and Blastx against NCBI human, mouse, and chicken UniGene databases. Nucleotide sequences matching with e values ≤ 1e−20 and protein sequences matching with e values ≤ 1e−10 were considered to be homologous with the query sequences. The UniGene IDs were then used to retrieve the relevant GO annotations. If multiple UniGene IDs were available, then we gave priority in the order of human/mouse/chicken to obtain a maximal number of clones with GO terms.
In Situ Hybridization.
In situ hybridization was performed as described in ref. 9. Briefly, brains of zebra finches or canaries were cut into 10-μm sagittal sections. Probes were labeled with33P-UTP and hybridized to fixed brain sections at 65°C overnight at 106cpm per slide. After washing, slides were exposed to x-ray film.
Supplementary Material
Acknowledgments
We thank B. Zhang, W. X. Zhang, and Dr. C. Zhao (all at the Genomics Resource Center, The Rockefeller University), and Dr. X. N. Wang (Department of Information Technology, The Rockefeller University) for their excellent support. This work was funded by National Institute of Health Grants DC 03492 (to X.L.) and MH18134 (to F.N.), and a grant from the Ellison Foundation (to F.N.). Parts of the bioinformatics work were supported by National Natural Science Foundation of China Grants 30621001 and 30570160 (to X.-J.W.).
Abbreviations
- CaM-TF
calmodulin-binding transcription factor
- FDR
false discovery rate
- HVC
high vocal center
- PCP-4
Purkinje cell protein 4.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) database (accession nos. EE049699–EE062017). Microarray data reported in this paper have been deposited in the Gene Expression Ominbus (GEO) database (accession no. GSE6412).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701619104/DC1.
References
- 1.Nottebohm F. Ann N Y Acad Sci. 2004;1016:628–658. doi: 10.1196/annals.1298.027. [DOI] [PubMed] [Google Scholar]
- 2.Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, Horita H, Patterson MA, White SA, Scharff C, et al. Proc Natl Acad Sci USA. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wade J, Peabody C, Coussens P, Tempelman RJ, Clayton DF, Liu L, Arnold AP, Agate R. J Neurosci Methods. 2004;138:199–206. doi: 10.1016/j.jneumeth.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Soares MB, Bonaldo MF, Jelene P, Su L, Lawton L, Efstratiadis A. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohner Anim Behav. 1990;39:369–374. [Google Scholar]
- 6.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Buylla A, Nottebohm F. Nature. 1988;335:353–354. doi: 10.1038/335353a0. [DOI] [PubMed] [Google Scholar]
- 8.Kirn JR, Fishman Y, Sasportas K, Alvarez-Buylla A, Nottebohm F. J Comp Neurol. 1999;411:487–494. [PubMed] [Google Scholar]
- 9.Li XC, Jarvis ED, Alvarez-Borda B, Lim DA, Nottebohm F. Proc Natl Acad Sci USA. 2000;97:8584–8589. doi: 10.1073/pnas.140222497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosik K, Krichevsky AM. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 13.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahnloser RH, Kozhevnikov AA, Fee MS. Nature. 2002;419:65–70. doi: 10.1038/nature00974. [DOI] [PubMed] [Google Scholar]
- 15.Nottebohm F, Stokes TM, Leonard CM. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- 16.Kirn JR, Nottebohm F. J Neurosci. 1993;13:1654–1663. doi: 10.1523/JNEUROSCI.13-04-01654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirn J, O'Loughlin B, Kasparian S, Nottebohm F. Proc Natl Acad Sci USA. 1994;91:7844–7848. doi: 10.1073/pnas.91.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wild JM, Williams MN, Howie GJ, Mooney R. J Comp Neurol. 2005;483:76–90. doi: 10.1002/cne.20403. [DOI] [PubMed] [Google Scholar]
- 20.Nottebohm F. PLoS Biol. 2005;3:e164. doi: 10.1371/journal.pbio.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tropea D, Kreiman G, Lyckman A, Mukherjee S, Yu H, Horng S, Sur M. Nat Neurosci. 2006;9:660–668. doi: 10.1038/nn1689. [DOI] [PubMed] [Google Scholar]
- 22.Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- 23.Suga Y, Jarnik M, Attar PS, Longley MA, Bundman D, Steven AC, Koch PJ, Roop DR. J Cell Biol. 2000;151:401–412. doi: 10.1083/jcb.151.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner TJ, Naef F, Nottebohm F. Science. 2005;308:1046–1049. doi: 10.1126/science.1108214. [DOI] [PubMed] [Google Scholar]
- 25.Nottebohm F. J Neurosci. 2002;22:624–628. doi: 10.1523/JNEUROSCI.22-03-00624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranscht B. J Cell Biol. 1988;107:1561–1573. doi: 10.1083/jcb.107.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutishauser U. J Cell Biol. 2000;149:757–760. doi: 10.1083/jcb.149.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez T, Morgan T, Davis N, Klin A, Morris A, Farhi A, Lifton RP, State MW. Am J Hum Genet. 2004;74:1286–1293. doi: 10.1086/421474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordeen KW, Nordeen EJ. Nature. 1988;334:149–151. doi: 10.1038/334149a0. [DOI] [PubMed] [Google Scholar]
- 30.Wilbrecht L, Williams H, Gangadhar N, Nottebohm F. J Neurosci. 2006;26:9135–9141. doi: 10.1523/JNEUROSCI.4869-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konishi M, Akutagawa E. Nature. 1985;315:145–147. doi: 10.1038/315145a0. [DOI] [PubMed] [Google Scholar]
- 32.Gaasterland T, Sensen CW. Biochimie. 1996;78:302–310. doi: 10.1016/0300-9084(96)84761-4. [DOI] [PubMed] [Google Scholar]
- 33.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith TF, Waterman MS. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 36.Kacharmina JE, Crino PB, Eberwine J. Methods Enzymol. 1999;303:3–18. doi: 10.1016/s0076-6879(99)03003-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.