Abstract
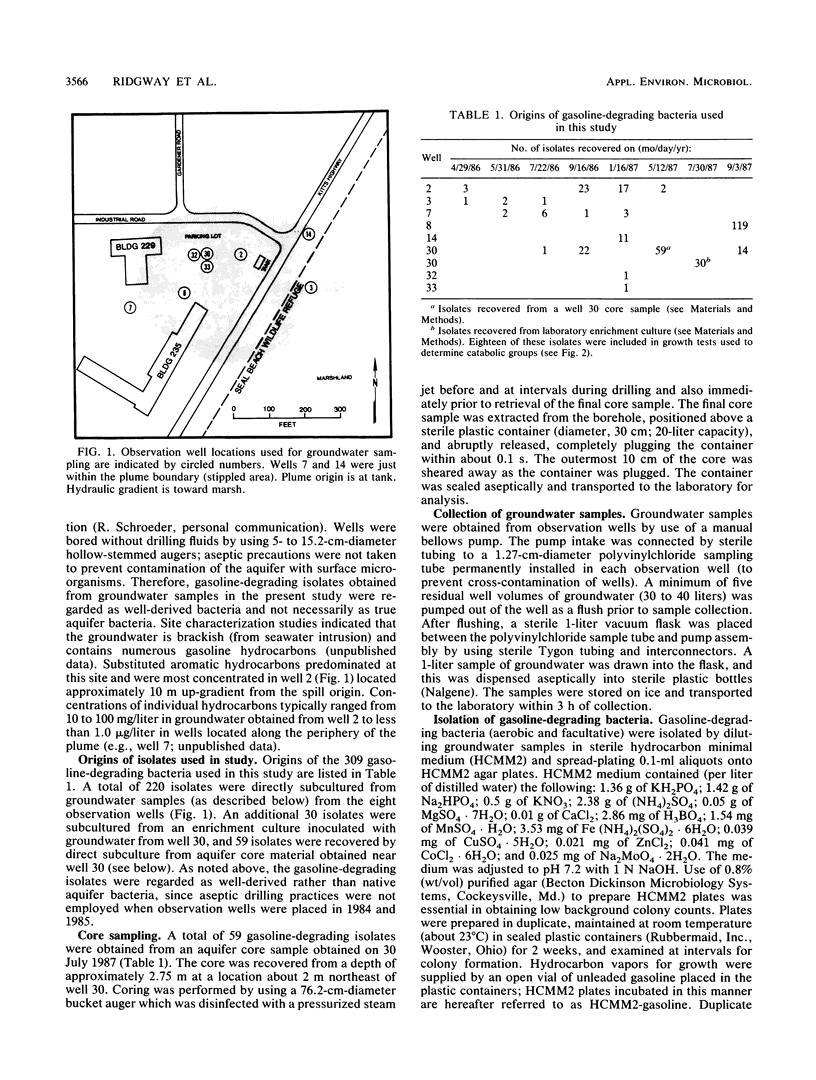
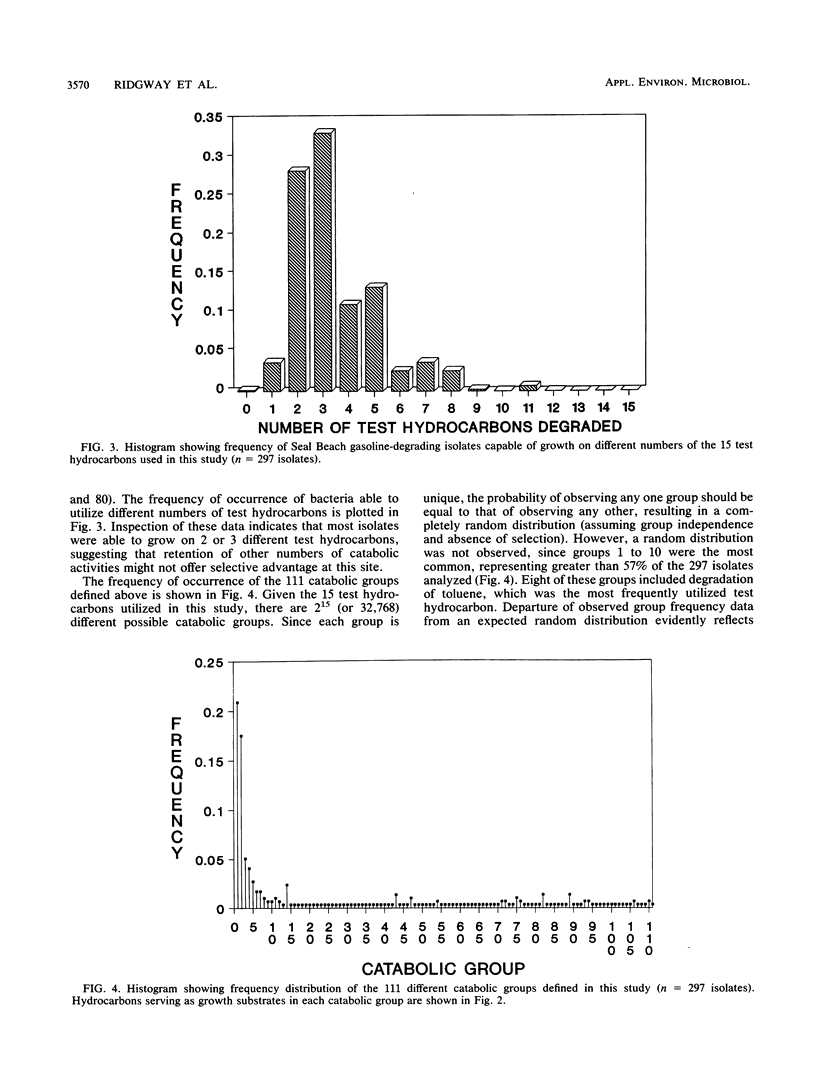
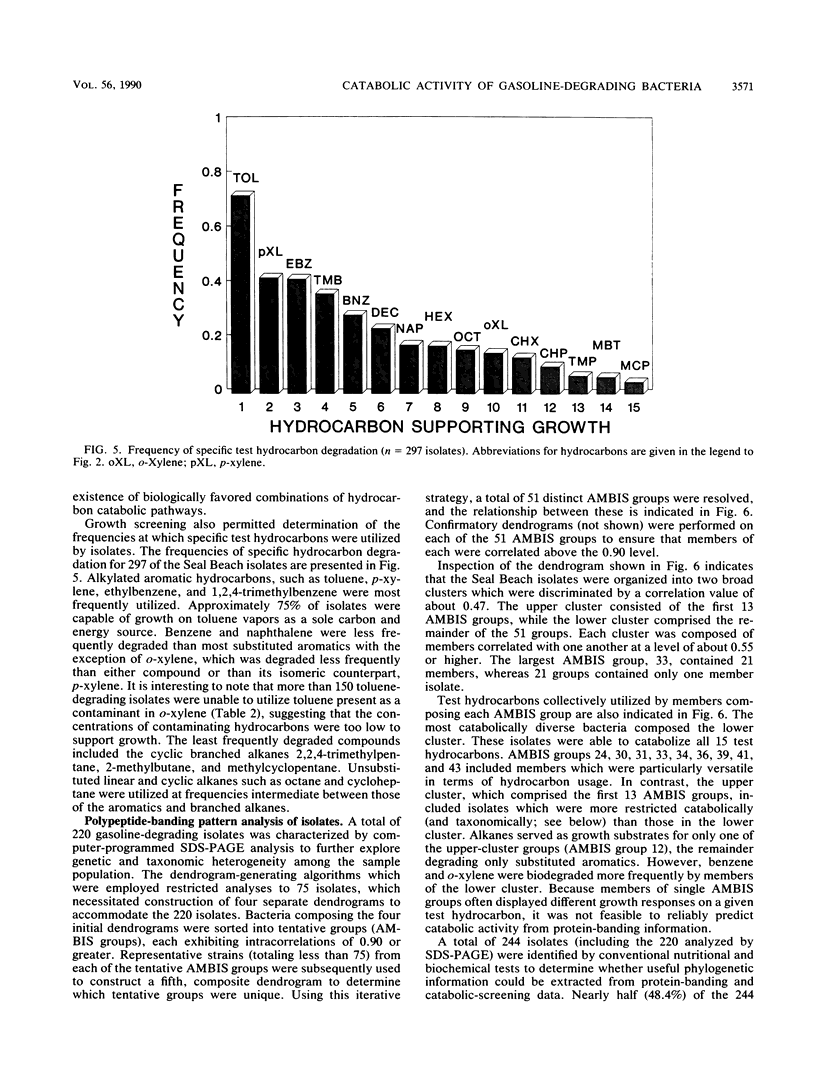
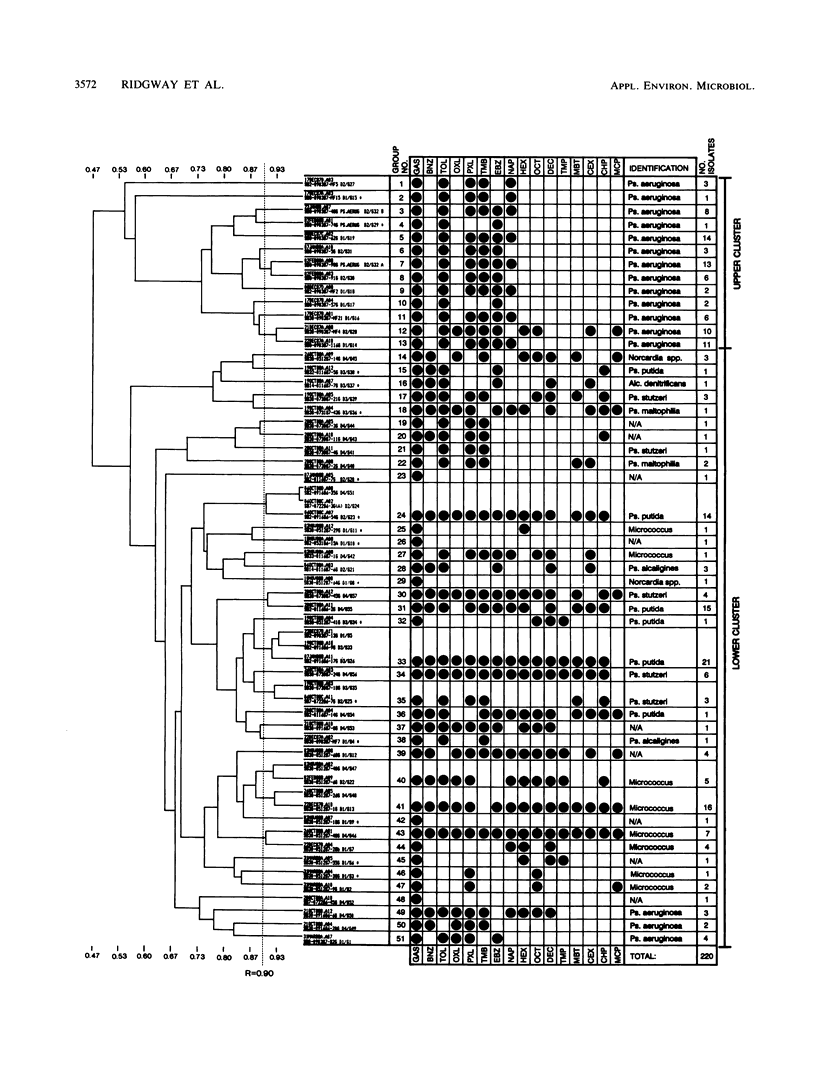
Approximately 300 gasoline-degrading bacteria were isolated from well water and core material from a shallow coastal aquifer contaminated with unleaded gasoline. Identification of 244 isolates revealed four genera: Pseudomonas, Alcaligenes, Nocardia, and Micrococcus, with pseudomonads making up 86.9% of bacteria identified. A total of 297 isolates was sorted into 111 catabolic groups on the basis of aerobic growth responses on 15 gasoline hydrocarbons. Each test hydrocarbon was degraded by at least one isolate. Toluene, p-xylene, ethylbenzene, and 1,2,4-trimethylbenzene were most frequently utilized as growth substrates, whereas cyclic and branched alkanes were least utilized. Most isolates were able to grow on 2 or 3 different hydrocarbons, and nearly 75% utilized toluene as a sole source of carbon and energy. Isolates were remarkably specific for hydrocarbon usage, often catabolizing only one of several closely related compounds. A subset of 220 isolates was sorted into 51 groups by polyacrylamide gel electrophoresis. Pseudomonas aeruginosa was partitioned into 16 protein-banding groups (i.e., subspecies) whose catabolic activities were largely restricted to substituted aromatics. Different members of subspecies groups defined by protein-banding pattern analysis often exhibited different growth responses on the same hydrocarbon, implying marked strain diversity. The catabolic activities of well-derived, gasoline-degrading bacteria associated with this contaminated aquifer are consonant with in situ adaptation at the site.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atlas R. M. Microbial degradation of petroleum hydrocarbons: an environmental perspective. Microbiol Rev. 1981 Mar;45(1):180–209. doi: 10.1128/mr.45.1.180-209.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill D. L., Ghiorse W. C. Characterization of subsurface bacteria associated with two shallow aquifers in oklahoma. Appl Environ Microbiol. 1985 Sep;50(3):580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D. F., Francis A. J., Bollag J. M. Microbial metabolism of homocyclic and heterocyclic aromatic compounds under anaerobic conditions. Microbiol Rev. 1987 Mar;51(1):43–59. doi: 10.1128/mr.51.1.43-59.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberger W. T., Jr Use of urea as a nitrogen fertilizer in bioreclamation of petroleum hydrocarbons in soil. Bull Environ Contam Toxicol. 1988 Jan;40(1):66–68. doi: 10.1007/BF01689388. [DOI] [PubMed] [Google Scholar]
- Ghiorse W. C., Wilson J. T. Microbial ecology of the terrestrial subsurface. Adv Appl Microbiol. 1988;33:107–172. doi: 10.1016/s0065-2164(08)70206-5. [DOI] [PubMed] [Google Scholar]
- Kersters K., De Ley J. Identification and grouping of bacteria by numerical analysis of their electrophoretic protein patterns. J Gen Microbiol. 1975 Apr;87(2):333–342. doi: 10.1099/00221287-87-2-333. [DOI] [PubMed] [Google Scholar]
- Perry J. J. Microbial cooxidations involving hydrocarbons. Microbiol Rev. 1979 Mar;43(1):59–72. doi: 10.1128/mr.43.1.59-72.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I. The AMBIS beta scanning system. Bioessays. 1985 Nov;3(5):225–229. doi: 10.1002/bies.950030510. [DOI] [PubMed] [Google Scholar]
- Swindoll C. M., Aelion C. M., Pfaender F. K. Influence of inorganic and organic nutrients on aerobic biodegradation and on the adaptation response of subsurface microbial communities. Appl Environ Microbiol. 1988 Jan;54(1):212–217. doi: 10.1128/aem.54.1.212-217.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]


