Abstract
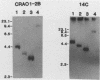
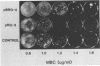
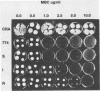
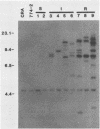
An altered beta-tubulin gene that confers resistance to benomyl [whose active ingredient is 2-(methoxycarbonylamino)benzimidazole (MBC)] was isolated from a DNA library of Aspergillus flavus and used as a selectable marker for transformation. The beta-tubulin gene was cloned into a plasmid vector containing the pyr-4 gene of Neurospora crassa, and transformants were selected either for uracil prototrophy or MBC resistance. Transformants selected for uracil prototrophy were of three phenotypic classes: sensitive, intermediate, and resistant to MBC. Transforming DNA appeared to integrate at several sites in the genome, with the more resistant phenotypes having more copies of the altered beta-tubulin gene than the sensitive and intermediate phenotypes. Transformants were also selected on medium containing MBC. The average frequency of transformation (1 to 3 transformants per micrograms of transforming DNA) was lower than that obtained by selection for uracil prototrophy, presumably because of failure to select transformants that contained few copies of the altered beta-tubulin gene. The sequence of the beta-tubulin gene was determined and compared with the published sequence of the benA gene of A. nidulans; the beta-tubulin gene was found to be highly conserved between the two Aspergillus species. Notable differences were that the beta-tubulin gene of A. flavus lacks intron 6 present in benA and has an additional leucine at position 148. This is the first gene sequence reported from an aflatoxin-producing fungus and adds to the growing body of knowledge of the beta-tubulin genes and their use as selectable markers for transformation of filamentous fungi.
Full text
PDF
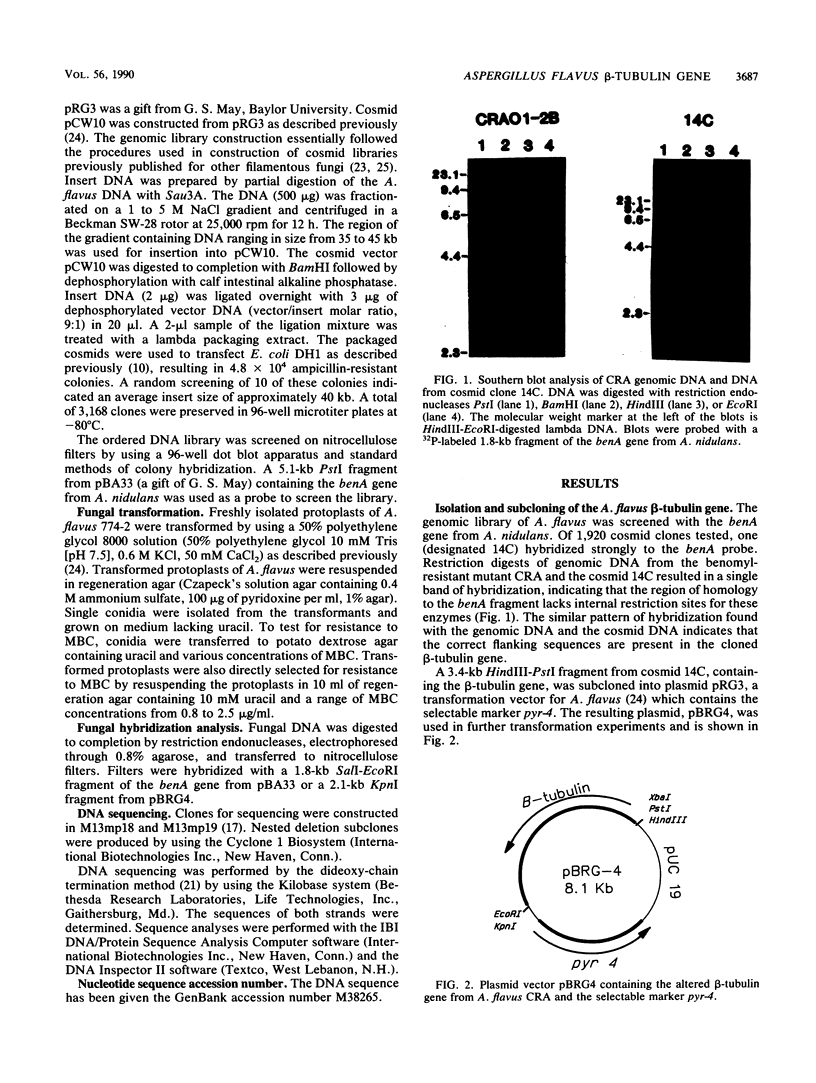





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballance D. J. Sequences important for gene expression in filamentous fungi. Yeast. 1986 Dec;2(4):229–236. doi: 10.1002/yea.320020404. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bhatnagar D., Ullah A. H., Cleveland T. E. Purification and characterization of a methyltransferase from Aspergillus parasiticus SRRC 163 involved in aflatoxin biosynthetic pathway. Prep Biochem. 1988;18(3):321–349. doi: 10.1080/00327488808062532. [DOI] [PubMed] [Google Scholar]
- Dunne P. W., Oakley B. R. Mitotic gene conversion, reciprocal recombination and gene replacement at the benA, beta-tubulin, locus of Aspergillus nidulans. Mol Gen Genet. 1988 Aug;213(2-3):339–345. doi: 10.1007/BF00339600. [DOI] [PubMed] [Google Scholar]
- Dutton M. F. Enzymes and aflatoxin biosynthesis. Microbiol Rev. 1988 Jun;52(2):274–295. doi: 10.1128/mr.52.2.274-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Mashaly R. I., Habib S. L., el-Deeb S. A., Salem M. H., Safwat M. M. Isolation, purification and characterization of enzyme(s) responsible for conversion of sterigmatocystin to aflatoxin B1. Z Lebensm Unters Forsch. 1988 Feb;186(2):118–124. doi: 10.1007/BF01042704. [DOI] [PubMed] [Google Scholar]
- May G. S., Gambino J., Weatherbee J. A., Morris N. R. Identification and functional analysis of beta-tubulin genes by site specific integrative transformation in Aspergillus nidulans. J Cell Biol. 1985 Sep;101(3):712–719. doi: 10.1083/jcb.101.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G. S., Tsang M. L., Smith H., Fidel S., Morris N. R. Aspergillus nidulans beta-tubulin genes are unusually divergent. Gene. 1987;55(2-3):231–243. doi: 10.1016/0378-1119(87)90283-6. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. G., Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Rinehart J. E., Mitchell B. L., Oakley C. E., Carmona C., Gray G. L., May G. S. Cloning, mapping and molecular analysis of the pyrG (orotidine-5'-phosphate decarboxylase) gene of Aspergillus nidulans. Gene. 1987;61(3):385–399. doi: 10.1016/0378-1119(87)90201-0. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa K. E. Genetics of Aspergillus flavus: linkage of aflatoxin mutants. Can J Microbiol. 1984 Jan;30(1):68–73. doi: 10.1139/m84-012. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshuk C. P., Seip E. R., Payne G. A., Adkins C. R. Genetic transformation system for the aflatoxin-producing fungus Aspergillus flavus. Appl Environ Microbiol. 1989 Jan;55(1):86–90. doi: 10.1128/aem.55.1.86-90.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton M. M., Timberlake W. E., Hondel C. A. A cosmid for selecting genes by complementation in Aspergillus nidulans: Selection of the developmentally regulated yA locus. Proc Natl Acad Sci U S A. 1985 Feb;82(3):834–838. doi: 10.1073/pnas.82.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir L. O., Hufford K. D. Precursor recognition by kinetic pulse-labeling in a toxigenic aflatoxin B1-producing strain of Aspergillus. Appl Environ Microbiol. 1981 Jul;42(1):168–173. doi: 10.1128/aem.42.1.168-173.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hartingsveldt W., Mattern I. E., van Zeijl C. M., Pouwels P. H., van den Hondel C. A. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet. 1987 Jan;206(1):71–75. doi: 10.1007/BF00326538. [DOI] [PubMed] [Google Scholar]