Abstract
Using comparative genomic hybridization (CGH) analysis, we, and others, have shown that there is a high and consistent incidence of chromosome 1q copy gain in human hepatocellular carcinoma (HCC). Chromosome 1 rearrangements, that involved peri-centromeric breakpoints, have also been frequently reported in karyotypic studies of HCC. Satellite DNA hypomethylation has been postulated as the mechanism underlying the induction of chromosome 1 peri-centromeric instability in many human cancers and in individuals with the rare recessive disorder ICF (immunodeficiency, centromeric heterochromatin instability, facial anomalies). In this study, we have investigated the role of DNA hypomethylation in 1q copy gain in HCC by examining the methylation status of chromosome 1 heterochromatin DNA (band 1q12). Thirty-six histologically confirmed samples of HCC were studied (24 paired tumor and adjacent nontumorous liver tissues, and 12 tumor only). Hypomethylation of satellite 2 (Sat2) DNA in 1q12 was analyzed by Southern blotting using methyl-sensitive enzyme digestion. In parallel, all cases were analyzed by CGH. A strong correlation between hypomethylated Sat2 sequences and 1q copy gain with a 1q12 breakpoint was found (P < 0.001). We postulate that such hypomethylation alters the interaction between the CpG-rich satellite DNA and chromatin proteins, resulting in heterochromatin decondensation, breakage and aberrant 1q formation. Spectral karyotyping further supported the presence of fragile 1q12 in HCC. Of particular interest was the finding of Sat2 DNA hypomethylation in 5 of 24 adjacent nontumorous liver tissues examined. These tissues showed no evidence of malignancy on histological examination nor did they display any CGH abnormalities. Our findings suggest a role for Sat2 demethylation in the early stages of the stepwise progression of liver carcinogenesis.
Hepatocellular carcinoma (HCC) is a highly malignant tumor that is prevalent in China, Southeast Asia, and sub-Saharan Africa. Associated genetic aberrations include common loss of heterozygosity on chromosomes 4q, 8p, 13q, and 16q, 1-3 and frequent gains of 8q, 17q, and 20q. 4-7 By comparative genomic hybridization (CGH), 8 we and others have identified a high incidence of 1q copy number gain in HCC (60 to 80%). 4-7 Karyotypic studies on HCC, on the other hand, indicated consistent structural abnormalities of chromosome 1. 9-14 In our recent spectral karyotyping (SKY) analysis on short-term cultured HCCs, unbalanced translocations of 1q that involved fusion at the centromeric heterochromatin region were found to be the most frequent karyotypic abnormality. 15 Current cytogenetic evidence therefore confers considerable importance of numerical 1q imbalance in liver carcinogenesis.
In cultures of normal human cell, DNA methylation inhibitors have been reported to induce peri-centromeric rearrangements of chromosome 1 at a very high frequency. 16,17 This may suggest that DNA demethylation activity is selective and preferentially targets the centromeric region of chromosome 1. Furthermore, classical satellite 2 (Sat2) hypomethylation has been detected in individuals with the rare recessive ICF syndrome (immunodeficiency, centromeric heterochromatin instability, facial anomalies), in which the chromosome 1 peri-centromeric rearrangements in mitogen-stimulated lymphocytes are characteristic. The Sat2 DNA is the major sequence of the unusually long heterochromatic region adjacent to the centromere of chromosome 1.
Several other human cancers also exhibit frequent chromosome 1 rearrangements that fuses in the vicinity of the heterochromatic region (1q12). The occurrence of chromosome 1q aberration in many different cancers suggests the likelihood of a common underlying mechanism. Hypomethylation of the constitutive heterochromatic region on chromosome 1 has been postulated as the cause for such chromosome 1 instability. Although the molecular mechanism that causes satellite DNA demethylation is still unclear, studies in breast cancer and Wilms’ tumor supported the hypothesis that common heterochromatin breakage on 1q12 is attributable to satellite hypomethylation and that this is likely to be the precursor to subsequent whole chromosome 1q translocations.
In this study, we investigated the role of heterochromatin DNA hypomethylation in the formation of aberrant 1q in HCC and its possible involvement in the stepwise progression of HCC development.
Materials and Methods
Patients
Tumorous liver tissues from 36 HCC patients (age 30 to 74 years; 75% male), who underwent surgical resection with curative intent and 24 paired adjacent nontumorous liver tissues were collected. Thirty-five patents were chronic carriers of viral hepatitis (97%), 32 cases of type B (HBV) and 3 cases of type C (HCV), with 81% arising from a cirrhotic liver. The disease stage of each case was classified according to the TNM staging criteria. 18 Of the 36 patients recruited, 3 cases (8%) were classified as stage I (T1N0M0), 27 (75%) as stage II (T2N0M0), 4 (11%) as stage III (T3N0M0), and 2 (6%) as stage IV (T4N0M0). An experienced liver pathologist confirmed the diagnosis of HCC and the nonmalignant status of adjacent liver tissues. The macroscopic and microscopic features of resected specimens were also reviewed for the presence or absence of underlying liver cirrhosis and the maximum diameter of each tumor was recorded.
Heterochromatin Hypomethylation by Southern Blot Analysis
Satellite DNA hypomethylation was examined by the Southern blot analysis described by Narayan and colleagues. 19 Two μg of DNA were digested in 20 U of CpG methyl-sensitive restriction enzyme BstBI (New England Biolabs, Beverly, MA) for 16 hours. Complete DNA digestion was indicated with the aid of an internal DNA control, λHindIII. Fractionated DNA blotted on Hybond N membrane (Amersham-Pharmacia, Arlington, Heights, IL) was probed against satellite 2 (Sat2), a major DNA component of the chromosome 1 heterochromatin. The Sat2 probe used was a single-stranded oligonucleotide of 18 mer that has a consensus sequence of 5′-TCGAGTCCATTCGATGAT-3′. Blotting hybridization in Rapid-Hyb buffer (Amersham-Pharmacia) was performed at 50°C using 5′-[γ-32P]-end radiolabeled dATP Sat2 probe. In each blot, normal liver and sperm DNAs were included as the methylated and hypomethylated standards, respectively. Posthybridization washes were performed in 5× sodium saline citrate/0.1% sodium dodecyl sulfate for 30 minutes at room temperature, followed by 1× sodium saline citrate/0.1% sodium dodecyl sulfate for 30 minutes at 50°C.
Using a phosphoimager (Instantimager; Packard, Australia), the approximate extent of hypomethylation in each lane was quantitated by comparing the ratio intensities of hybridized fragments <4 kb to those >4 kb molecular weight. DNA samples with ratios <0.7 were considered to have a normal level of Sat2 methylation, whereas those with values >1.1 were considered to be extensively hypomethylated. Ratio values between 0.7 to 1.1 were considered to display a moderate level of hypomethylation. The cut off value of 0.7 for the presence of hypomethylation was assigned based on the degree of methylation obtained from four normal liver tissues (mean plus 1 SD). These tissues were neither cirrhotic nor viral infected, and had no apparent malignant morphology on histological examination. The ratio of 1.1 for extensive hypomethylation was established from four positive sperm controls (mean minus 1 SD). Ratio values for moderate hypomethylation were those that were between the normal level of methylation and extensive hypomethylation. Statistical analysis for the association between 1q copy gain and Sat2 hypomethylation was performed by the Fisher’s exact test.
Comparative Genomic Hybridization
The CGH protocol was performed according to the method of Kallioniemi and colleagues 8 with modifications described in Wong and colleagues. 5 Briefly, differentially labeled tumor and normal DNA with biotin-16-dUTP (Boehringer Mannheim, Mannheim, Germany) and dig-11-dUTP (Boehringer Mannheim) were co-hybridized onto normal metaphase chromosomes. After hybridization, biotin signals were detected through avidin conjugated-fluorescein isothiocyanate antibody (Sigma, St. Louis, MO), and dig-labeled DNA visualized by tetramethylrhodamine isothiocyanate-conjugated antibody (Sigma). Chromosomes counterstained with 4′,6-diamidino-2-phenylindole (DAPI) were captured through a cooled charge-coupled device camera mounted on a Leitz DM RB (Leica, Wetzlar, Germany) fluorescence microscope. Three band-pass filter sets (DAPI, fluorescein isothiocyanate, and tetramethylrhodamine isothiocyanate) arranged in an automated filter-wheel were used for image acquisition. CGH software ver 3.1 on Cytovision (Applied Imaging Ltd., Sunderland, UK) was used for digital image analysis of fluorescence intensity. Average ratio profiles of 10 to 12 metaphases were calculated based on chromosome identification of the inverted DAPI. 3 Thresholds for gains and losses were defined as the theoretical value of 1.25 and 0.75, respectively.
Spectral Karyotyping
SKY analysis was performed on the aberrant metaphases of cases H25, H26, H29, H32, and H34. The short-term culture of primary tumors was performed according to the procedure described in Wong and colleagues, 15 and subsequent SKY analysis according to the method described by Schröck and colleagues. 20 Briefly, tumorous liver tissues digested by collagenase (type II) were seeded in RPMI 1640 medium supplemented with 16% fetal bovine serum, 35 U/ml penicillin, 35 μg/ml streptomycin, 10 ng/ml selenium, 10 μg/ml transferrin, and 10 μg/ml insulin. At 80% confluency, which took 3 to 5 days, cells were harvested for metaphase chromosomes by colchicine. Labeled SKY probe mixture (Applied Spectral Imaging Ltd., Migdal Haemek, Israel) was applied onto the tumor metaphases. After posthybridization washes, indirectly labeled probes were visualized using fluorescence-conjugated antibodies. Chromosomes counterstained in DAPI was acquired using a SD200 Spectracube (Applied Spectral Imaging,) mounted on a Leica DMRXA microscope (Leica). Spectral information obtained on each chromosome was analyzed by the SkyView software ver 1.6.
Results
Heterochromatin Hypomethylation and 1q Gain
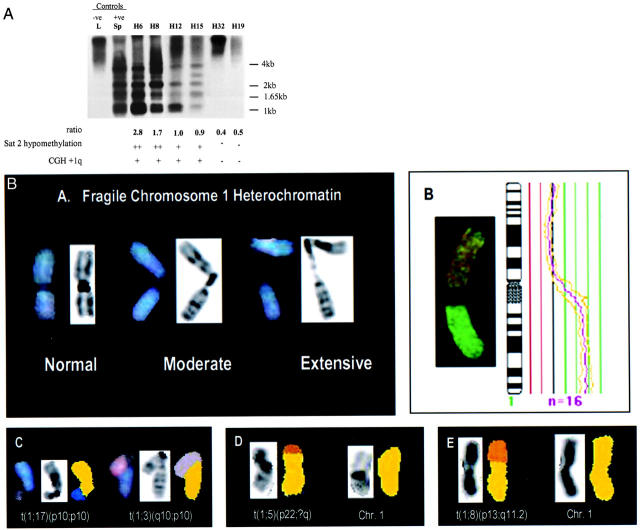
Sat2 DNA hypomethylation was detected in 25 of 36 HCC tissues studied (69%) (Table 1) ▶ . Examples of hypomethylated Sat2 cases are shown in Figure 1a ▶ . Twenty-two of the 25 cases with Sat2 hypomethylation displayed 1q copy number gain as suggested from CGH analysis (88%). Hybridized chromosome from CGH indicated a consistent breakpoint within the heterochromatic region, band 1q12, in these 22 cases (Figure 1b) ▶ . Although chromosome 1 instability was not suggested in the remaining three cases (H9, H27, and H29), the finding of extensive heterochromatin hypomethylation prompted us to undertake further karyotypic investigation (Table 1) ▶ . SKY analysis on H29 indicated an unbalanced translocation of chromosome 1p and 1q, t(1;17)(p10;p10) and t(1;3)(q10;p10), respectively (Figure 1b) ▶ , suggesting no net gain or loss of chromosome 1 material. Of particular interest, SKY analysis in cases H25, H26, and H29 indicated copies of chromosome 1 with much extended heterochromatic region, suggesting fragility of the 1q12 segment (Figure 1b) ▶ .
Table 1.
Heterochromatin Hypomethylation and CGH Finding on HCC and Adjacent Nontumorous Liver Tissues
| Case no. | Age/sex | Viral status | Cirrhosis | Tumor diameter (cm) | TNM stage | CGH analysis | Sat2 hypomethylation | ||
|---|---|---|---|---|---|---|---|---|---|
| Tumor | Adjacent tissue | Tumor | Adjacent tissue† | ||||||
| H1 | 69 /M | HBV | − | 11.0 | T2N0M0 | +1q, +2,−4,+5q31-qter,+6pter-p22,−9,+10p,+14q,−15q,+16, −17pter-p13,+17q11-q21,+17q22-qter,+20, −21q | Balanced | ++ | + |
| H2 | 37 /M | HBV | − | 6.9 | T2N0M0 | +1q, +2q21-q24,+3p,+4p,−4q,+5,−6q22-q26,+7p22-p11.2,+7q21-qter, −8pter-p12,+8p11.2-qter,−9pter-p21,+9p13-p12,+10q,−11q14-qter, −12pter-p12,+12p11.2-q22,−13q,−14q,−17p,+18p,−18q21-qter, +Y,+Xq13-qter | Balanced | ++ | + |
| H3 | 59 /M | HBV | + | 4.0 | T3N0M0 | −1p36, +1q,+3pter-p13,−4q13-qter,+6pter-p22,+8q,−12q24-qter, −16q13,−16q24 | Balanced | + | + |
| H4 | 58 /M | HBV | − | 4.5 | T2N0M0 | +1q12-q32, +3q12-q23,−4q,−5q,+8q,+10pter-q21,−11q21-qter,−13q12-q21, +13q32,+14q11.2,+17p12-q21,+18q11.2,+19p13-q13.2, +20q12-q13.2 | Balanced | + | + |
| H5 | 58 /F | HBV | − | 7.0 | T2N0M0 | −1p35-p21, +1q,−3p,−4q12-q21,−6p12-qter,−9q13-q34,+11q12-q13, −13q,−14q22-qter,+15q,+20,+21q,−Y,+X | Balanced | + | + |
| H6 | 30 /F | HBV | + | 8.0 | T2N0M0 | +1q, −2q32-qter,−4q28-qter,−7q32-qter,−8p,+8q,+9q32-qter,+10p, −10q,−11q,−12p,−13q12-q31,−14q,−16q,+16pter-p11.2, −Xpter-q22 | Balanced | ++ | − |
| H7 | 74 /F | HCV | + | 3.0 | T2N0M0 | +1q, +11q12-q14 | Balanced | ++ | − |
| H8 | 69 /F | HBV | + | 8.0 | T2N0M0 | −1pter-p35, +1q,+2pter-p21,−4q,−5q,+7pter-q33,+7q21-q31, −9pter-p13,−12p,+13q22-qter,−16q, −Xpter-p21 | Balanced | ++ | − |
| H9 | 51 /M | HBV | + | 7.0 | T2N0M0 | +7p, −8p22-p12,+8p11-qter,+11pter-p14,−14q,−15q,−17pter-q11, +17q12-qter,−Y | Balanced | ++ | − |
| H10 | 61 /M | HCV | + | 2.5 | T2N0M0 | +1q, −2q11-q32,−4q13-qter,+5q31-qter,+6p,−6q12-q22,−8p, +10pter-p12,−16q,+17q22-qter,+20q11.2-q13.2, +Xq21-q27 | Balanced | ++ | − |
| H11 | 51 /M | HCV | + | 4.4 | T2N0M0 | +1q12-q42, +8q,+Xq | Balanced | ++ | − |
| H12 | 55 /M | HBV | + | 5.8 | T3N0M0 | +1q, +3q22-qter,−4q,−5q,+6p,−8p,−14q22-qter | Balanced | + | − |
| H13 | 55 /M | HBV | + | 7.0 | T2N0M0 | +1q, −4,+5pter-q33,+8q,+10pter-q21,−13q21-qter,−14q21-qter,−16q +18q21-qter,+20p,+20q,+X | Balanced | + | − |
| H14 | 51 /M | N/A | + | 3.5 | T2N0M0 | +1q, +5q15-qter,+7,−8pter-p12,+8q21-qter,−10q23-qter,−16q,−21q, −22q,+X | Balanced | + | − |
| H15 | 61 /F | HBV | + | 11.0 | T2N0M0 | +1q, +8q13-qter | Balanced | + | − |
| H16 | 69 /M | HBV | + | 2.0 | T1N0M0 | +1q, +5q14-qter,−8p21-q13,+8q14-qter,−17p,−22q | Balanced | + | − |
| H17 | 48 /M | HBV | − | 7.0 | T2N0M0 | +1q12-q25, +2pter-p12,−4q21-q28,−10q25-qter,−13q14-q21, +X | Balanced | + | − |
| H18 | 47 /M | HBV | + | 3.9 | T2N0M0 | +1q, −6q24-qter,+12q15-q24.1,−13q,−15q13-qter,−17p,+18q12-q21, −19p,−22q,+X | Balanced | + | − |
| H19 | 51 /M | HBV | + | 8.5 | T2N0M0 | −4q12-q32, −5q11-q14,+6q25-qter,+8q22-qter,−10p11-qter,−11q14-qter, +12q12-q21,−12q23-qter,−13q,−16q,−17p,+19q,−21q, +Xq22-qter | Balanced | − | − |
| H20 | 34 /F | HBV | + | 3.2 | T2N0M0 | +1q25-qter, +5,−6q12-q24,+7q21-q31,−8pter-q21.3,−9pter-p22,+12q24.1, +15q23-qter,−17p11.2-q24,−18q | Balanced | − | − |
| H21 | 52 /M | HBV | + | 4.9 | T4N0M0 | +7, −8p,+8q | Balanced | − | − |
| H22 | 44 /F | HBV | − | 2.7 | T2N0M0 | −8p, +8q,−9pter-p22,−13q,−16q,−17p13,−18, +Xq | Balanced | − | − |
| H23 | 54 /M | HBV | + | 2.7 | T2N0M0 | −1pter-p21, +1p21-qter,+4p,−4q,+6pter-q14,−6q15-qter,−8pter-p21, +8p12-qter,−16 | Balanced | − | − |
| H24 | 60 /M | HBV | + | 2.4 | T3N0M0 | +1p13-qter, −4,+7q21-qter,−8p,+8q,−12q21-qter,−13q21-qter,−16q,−17p13, −19q,−20q13,−22q | Balanced | − | − |
| H25 | 67 /F | HBV | + | 3.0 | T2N0M0 | +1q, +2q32-qter,+6pter-p21,+7,+17q,+Xq | N/A | ++ | N/A |
| H26 | 60 /M | HBV | − | 11.0 | T2N0M0 | +1q, +5q13-qter,−8p,+8q,+10p,−13q12-q22,+13q22-q34,−16q, +X | N/A | ++ | N/A |
| H27 | 33 /M | HBV | + | 2.0 | T1N0M0 | −4pter-p14, −4q,+5,+6pter-p21.1,+8pter-q23,−11pter-p12,+12q13-q22, −13q12-q22,+14q13-q22,−16q,−17p,+18p,−18q,−19p, −20pter-p11.2,−22q | N/A | ++ | N/A |
| H28 | 69 /M | HBV | + | 6.5 | T2N0M0 | +1q, −4,+6p,−8p21-p22,−9,+16p,−16q,+17pter-q11.2,+18p11.2-qter, +19q13.1,−19q13.2-qter,−Y,+Xq22-qter | N/A | ++ | N/A |
| H29 | 40 /M | HBV | + | 5.5 | T4N0M0 | −8pter-p21, −8p12-qter,+10p,+12q,+17q,+Xpter-p21 | N/A | ++ | N/A |
| H30 | 74 /F | HBV | + | 3.6 | T2N0M0 | +1q, +5p,+8q | N/A | + | N/A |
| H31 | 60 /M | HBV | + | 2.6 | T2N0M0 | −1p, +2q21-qter,−6q13-qter,+8q | N/A | + | N/A |
| H32 | 39 /M | HBV | + | 8.5 | T2N0M0 | +1p13-qter, +2q14.1-q33,+5p,+8q11.2-q22,+11q14-qter,−16p | N/A | − | N/A |
| H33 | 68 /M | HBV | + | 4.8 | T2N0M0 | −4p21-qter, +6q24-qter,−11q22-qter,+17q23-qter | N/A | − | N/A |
| H34 | 65 /M | HBV | + | 1.4 | T1N0M0 | −1p22-pter, +3,+4p,−4q24-qter,+5p,−5q,−8p,+11,+12,−13q12-q21, +13q22-qter,−14q21-qter,−15q22-qter,−16,−17p,−18q,+19,+20, −21q,+22q | N/A | − | N/A |
| H35 | 57 /M | HBV | + | 4.0 | T3N0M0 | +1p21-qter, +8q21.2-qter,+13q22-qter | N/A | − | N/A |
| H36 | 71 /M | HBV | + | 6.5 | T2N0M0 | +1q, +6p,−14q13-qter | N/A | − | N/A |
Thirty-six tumor samples and 24 paired adjacent nontumorous liver tissues were studied by CGH and analyzed for Sat2 hypomethylation. ++, Indicates extensive hypomethylation of the Sat2 DNA in the heterochromatic region of chromosome 1; +, indicates moderate Sat2 hypomethylation; and −, indicates absence of Sat2 hypomethylation.
†Moderate hypomethylation with ratio values ranging from 0.7 to 0.9 was detected in five adjacent nontumorous livers (H1 to H5).
Genetic changes detected by CGH are listed as gains and losses. Chromosome 1 imbalances are highlighted in bold and underlined.
HBV, hepatitis B surface antigen positive; HCV, anti-hepatitis C positive; N/A, not determined.
Figure 1.
Top: Methylation status of chromosome 1 heterochromatin DNA in HCC. Hypomethylation status of satellite 2 (Sat2) DNA of chromosome 1 was studied by Southern blot. DNA extracted from HCC was digested by methyl-sensitive enzyme BstB1 and probed against Sat2. Liver (L) and sperm (Sp) DNAs were used as the methylated (negative control) and hypomethylated (positive control) standards, respectively. The degree of hypomethylation in each lane was quantitated by comparing the ratio intensities of hybridized fragments smaller than 4 kb to those larger than 4-kb molecular weight. DNA samples with ratios <0.7, 0.7 to 1.1, and >1.1 were considered to have a normal level of methylation, moderate level of Sat2 hypomethylation, and extensive hypomethylation, respectively. Bottom: Molecular cytogenetic analysis on HCC. A: SKY analysis on H29 revealed copies of chromosome 1 with moderate to extensive decondensation of the heterochromatic region. Repetitive DNA sequences within the heterochromatic region had been suppressed by Cot-1 DNA but the reverse DAPI image indicated fragility of the segment. B: CGH analysis on HCC revealed frequent 1q copy number gain. Visual inspection of hybridized chromosome often indicated a strong staining green region of tumor DNA on the q-arm and a suggested breakpoint within 1q12. The figure shown illustrates the 1q copy gain detected in case H2. Fluorescent ratio intensities is plotted alongside the chromosome ideogram and the ratio profile of 16 chromosomes (n = 16, pink line) is depicted with 95% confidence interval (gold lines). C: SKY analysis on H29 supported CGH finding for the presence of balanced chromosome 1. The finding of chromosome 1p and 1q translocation, t(1;17)(p10;p10) and t(1;3)(q10;p10), suggested no net gain or loss of chromosome 1 material. D: Chromosome 1 translocation in case H34. Classified image is displayed alongside the reverse DAPI image. SKY analysis indicated a translocation between chromosomes 1 and 5, t(1;5)(p22;?q), with a p-arm breakpoint on band 1p22. E: Classified karyotype of chromosome 1 in case H32. Reverse DAPI-banding pattern indicated a translocation between chromosomes 1 and 8, t(1;8)(p13;q11.2), with a chromosome 1 breakpoint on p13.
In 11 HCC cases with a normal level of Sat2 methylation, 4 showed no evidence of an unbalanced chromosome 1 (H19, H21, H22, and H33). Gain on chromosome 1 was detected in six cases (H20, H23, H24, H32, H35, and H36), and one case exhibited loss of regional 1p (H34). All but one case displayed a breakpoint outside the region of 1q12 (Table 1) ▶ . The assignment of a breakpoint was further supported by SKY analysis, which indicated a p-arm breakage in cases H32 and H34 (Figure 1b) ▶ . A 3×2 contingency statistical analysis was performed to examine the correlation between 1q gain, Sat2 hypomethylation, and the significance of the 1q12 breakpoint (Table 2) ▶ . Fisher’s exact test indicated a strong correlation between heterochromatin DNA hypomethylation and 1q copy gain with a breakpoint at 1q12 (P < 0.001). In 36 HCC tissues studied, no obvious relation between the degree of Sat2 hypomethylation and clinical staging, tumor size, or viral infection could be discerned.
Table 2.
A Correlation between Heterochromatin Hypomethylation and 1q12 Breakpoint in 1q Copy Number Gain
| Chromosome 1 heterochromatin hypomethylation | ||
|---|---|---|
| Positive | Negative | |
| Breakpoint within 1q12 with evidence of 1q gain | 22 | 1 |
| Breakpoint outside 1q12 with evidence of 1q gain | 0 | 6 |
| Absence of chromosome 1q12 breakpoint with no evidence of 1q gain | 3 | 4 |
Fisher’s exact test, P < 0.001.
Heterochromatin Hypomethylation in Adjacent Hepatitis-Infected Liver Tissues
Five of 24 adjacent liver tissues studied displayed a moderate level of Sat2 hypomethylation (H1 to H5). All cases were viral hepatitis B related, and arose from a noncirrhotic liver except for case H3. Histological examination revealed no apparent malignant phenotype in these tissues, and CGH analysis did not indicate genomic imbalances in any of the 24 adjacent liver tissues examined (Table 1) ▶ .
Discussion
Methylation of CpG dinucleotides functions to maintain the stability of chromosome structures. 21 Satellite DNA in the heterochromatin, in particular, is more heavily methylated at the CpG islands than the rest of the genomic DNA. 22,23 It has been postulated that under-methylated satellite sequences confer abnormal chromatin structures and predispose to chromosomal instability by enhancing chromosome recombinations. 24 A high frequency of chromosome rearrangements with peri-centromeric or heterochromatic breakpoints has been observed in many tumors, which may suggest a relationship between satellite hypomethylation, chromosome instability and carcinogenesis. 25,26
Our current finding supported a relationship between hypomethylated Sat2 sequences and recurrent aberrant 1q formation. We investigated the methylation status of Sat2 DNA, rather than the centromeric satellite (Sat-α), as Sat2 is a more CpG rich region than Sat-α (AT-rich region). In this series, methyl-sensitive endonuclease analysis showed a reduced methylation of classical Sat2 in 76% of HCC cases that displayed 1q copy number gain. In particular, we found Sat2 hypomethylation to be strongly associated with a 1q12 breakpoint (P < 0.001) (Table 2) ▶ . In our recent karyotypic study on human HCCs, SKY analysis did not identify nonrandom chromosome 1 rearrangements, but rather frequent unbalanced 1q translocations with breakage in the vicinity of 1q12. Consistent localization of breakpoints within the heterochromatic region in HCC therefore suggests an important pathogenic consequence of Sat2 hypomethylation in 1q abnormalities. Structural decondensation of 1q12 is likely to result in centromeric fragility, somatic pairing, and the formation of jumping 1q translocations. We were able to support the presence of 1q12 segment decondensation by the finding of a fragile heterochromatic region in three cases (H25, H26, and H29) that displayed extensive Sat2 hypomethylation (Figure 1b) ▶ .
DNA methylation patterns are often altered in cancer. In a number of human malignancies, regional hypermethylation of the promoter region of critical tumor suppressor gene(s) results in silencing of transcriptional activity, and global DNA hypomethylation leading to activation of proto-oncogenes and re-expression of provirus sequences has been described. 27,28 Given the multistep nature of liver carcinogenesis, cancer-associated genetic and epigenetic alterations are probable in the putative precancerous liver lesions, the surrounding viral hepatitis-infected cirrhotic tissues. Indeed, microsatellite instability and aberrant DNA methylation of E-cadherin, p16, and c-myc have been reported in the noncancerous liver tissues of HCC. 29-32 In our current series, 24 adjacent nonmalignant liver tissues had been examined for heterochromatin hypomethylation. Similar to our previous report 5 and that of a recent study from Taiwan, 33 CGH aberrations were not found in any of the adjacent liver tissues. However, a moderate level of Sat2 demethylation was detected in 20% of the viral hepatitis B-related surrounding liver. A viral origin in the induction of peri-centromeric fragility in human neoplasms has been previously suggested. 34,35 Although the role of hepatitis B infection, a DNA virus, in the demethylation of repeat sequences is unclear, gene products of cancer-associated DNA virus, such as SV40 and HPV, are known to alter cellular proteins and affect cell-cycle checkpoints, thereby inducing karyotypic instability. 36
Genome-wide hypomethylation facilitates tumor progression. Demethylation of the repetitive sequences, such as LINE1, alphoid repeats, and Alu, constitute a major part of the global hypomethylation in tumor development. Although LINE1 hypomethylation has not been suggested in surrounding liver tissues of HCC, 37 methylationsensitive representational differential analysis 38 has indicated that global hypomethylation is not homogenous throughout the entire genome. Instead, hypomethylatedregions are scattered in the genome. Our present finding therefore suggests that heterochromatin demethylation precedes genome-wide hypomethylation, whereby heterochromatin fragility results in the clonal evolution of cells with extra copies of 1q. Chromosome 1q copy number gain may confer proliferative advantages that contribute to the natural evolution of HCC progression.
Footnotes
Address reprint requests to Nathalie Wong, Department of Clinical Oncology, The Chinese University of Hong Kong, Prince of Wales Hospital, Shatin, N. T., SAR Hong Kong, China. E-mail: natwong@cuhk.edu.hk.
This work was supported by The Kadoorie Charitable Foundations (to the Hong Kong Cancer Genetics Research Group), Hong Kong; the Research Grants Council of the Hong Kong Special Administrative Region; and the Providence Foundation Limited, Hong Kong.
References
- 1.Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Harada H, Yagura M, Matsubara K, Nakamura Y: Allelotype study of primary hepatocellular carcinoma. Cancer Res 1991, 51:89-93 [PubMed] [Google Scholar]
- 2.Nagai H, Pineau P, Tiollais P, Buendia MA, Dejean A: Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene 1997, 14:2927-2933 [DOI] [PubMed] [Google Scholar]
- 3.Boige V, Laurent-Puig P, Fouchet P, Flejou JF, Monges G, Bedossa P, Bioulas S, Capron F, Schmitz A, Olschwang S, Thomas G: Concerted nonsyntenic allellic losses in hyperploid hepatocellular carcinoma as determined by a high-resolution allelotype. Cancer Res 1997, 57:1986-1990 [PubMed] [Google Scholar]
- 4.Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A: Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosom Cancer 1997, 18:59-65 [PubMed] [Google Scholar]
- 5.Wong N, Lai P, Lee S-W, Fan S, Pang E, Liew C-T, Sheng Z, Lau JW, Johnson PJ: Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis: relationship to disease stage, tumor size, and cirrhosis. Am J Pathol 1999, 154:37-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K: Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology 1999, 29:1858-1862 [DOI] [PubMed] [Google Scholar]
- 7.Qin LX, Tang ZY, Sham JS, Ma ZC, Ye SL, Zhou XD, Wu ZQ, Trent JM, Guan XY: The association of chromosome 8p deletion and tumor metastasis in human hepatocellular carcinoma. Cancer Res 1999, 59:5662-5665 [PubMed] [Google Scholar]
- 8.Kallioniemi A, Kallioniemi O-P, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 9.Simon D, Munoz SJ, Maddrey WC, Knowles BB: Chromosomal rearrangements in a primary hepatocellular carcinoma. Cancer Genet Cytogenet 1990, 45:225-260 [DOI] [PubMed] [Google Scholar]
- 10.Bardi G, Johansson B, Pandis N, Heim S, Mandahl N, Andren-Sandberg A, Hagerstrand I, Mitelman F: Cytogenetic findings in three primary hepatocellular carcinomas. Cancer Genet Cytogenet 1992, 58:191-195 [DOI] [PubMed] [Google Scholar]
- 11.Bardi G, Rizou H, Michailakis E, Dietrich C, Pandis N, Heim S: Cytogenetic findings in three primary hepatocellular carcinomas. Cancer Genet Cytogenet 1998, 104:165-166 [DOI] [PubMed] [Google Scholar]
- 12.Werner M, Nolte M, Gergii M, Klempnauer J: Chromosome 1 abnormalities in hepatocellular carcinoma. Cancer Genet Cytogenet 1993, 66:130. [DOI] [PubMed] [Google Scholar]
- 13.Chen HL, Chen YC, Chen DS: Chromosome 1p aberrations are frequent in human primary hepatocellular carcinoma. Cancer Genet Cytogenet 1996, 86:102-106 [DOI] [PubMed] [Google Scholar]
- 14.Parada LA, Hallen M, Transberg KG, Hagerstarnd I, Bondeson L, Mitelman F, Johansson B: Frequent rearrangements of chromosomes 1, 7, and 8 in primary liver cancer. Genes Chromosom Cancer 1998, 23:26-35 [DOI] [PubMed] [Google Scholar]
- 15.Wong N, Lai P, Pang E, Leung T W-T, Lau J W-Y, Johnson PJ: A comprehensive karyotypic study on human hepatocellular carcinoma by spectral karyotyping. Hepatology 2000, 32:1060-1068 [DOI] [PubMed] [Google Scholar]
- 16.Hernandez R, Frady A, Zhang XY, Varela M, Ehrlich M: Preferential induction of chromosome 1 multibranched figures and whole-arm deletions in a human pro-B cell line treated with 5-azacytidine or 5-azadeoxycytidine. Cytogenet Cell Genet 1997, 76:196-201 [DOI] [PubMed] [Google Scholar]
- 17.Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, Varela M, Ehrlich M: DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res 1997, 379:33-41 [DOI] [PubMed] [Google Scholar]
- 18.Beahrs OH, Henson DE, Hutter RVP, Kennedy BJ (eds). Handbook for Staging of Cancer. Philadelphia, J. B. Lippincott, 1993
- 19.Narayan A, Ji W, Zhang X-Y, Marrogi A, Graff JR, Baylin SB, Ehrlich M: Hypomethylation of peri-centromeric DNA in breast adenocarcinoma. Int J Cancer 1998, 77:833-838 [DOI] [PubMed] [Google Scholar]
- 20.Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith M, Ning Y, Ledbetter DH, Bar-Am I, Soenksen Y, Garini T, Ried T: Multicolor spectral karyotyping of human chromosomes. Science 1996, 273:494-496 [DOI] [PubMed] [Google Scholar]
- 21.Hori TA: Induction of chromosome decondensation, sister-chromatid exchanges and endoreduplications by 5-azacytidine, an inhibitor of DNA methylation. Mutat Res 1983, 121:47-52 [DOI] [PubMed] [Google Scholar]
- 22.Ehrlich M, Wang RY: 5-Methylcytosine in eukaryotic DNA. Science 1981, 212:1350-1357 [DOI] [PubMed] [Google Scholar]
- 23.Feinstein SI, Racaniello VR, Ehrlich M, Gehrke CW, Miller DA, Miller OJ: Pattern of undermethylation of the major satellite DNA of mouse sperm. Nucleic Acids Res 1985, 13:3969-3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engler P, Haasch D, Pinkert CA, Doglio L, Glymour M, Brinster R, Storb U: A strain-specific modifier on mouse chromosome 4 controls the methylation of independent transgene loci. Cell 1991, 65:939-947 [DOI] [PubMed] [Google Scholar]
- 25.Sawyer JR, Tricot G, Mattox S, Jagannath S, Barlogie B: Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood 1998, 91:1732-1741 [PubMed] [Google Scholar]
- 26.Qu G, Dubeau L, Narayan A, Yu MC, Ehrlich M: Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mutat Res 1999, 423:91-101 [DOI] [PubMed] [Google Scholar]
- 27.Fearon ER, Vogelstein BA: Genetic model for colorectal tumorigenesis. Cell 1990, 61:759-767 [DOI] [PubMed] [Google Scholar]
- 28.Baylin SB, Makos M, Wu J, Chiu Yen RW, de Bustros A, Vertino P, Nelkin BD: Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells 1991, 3:383-390 [PubMed] [Google Scholar]
- 29.Aiba N, Nambu S, Inoue K, Sasaki H: Hypomethylation of the c-myc oncogene in liver cirrhosis and chronic hepatitis. Gastroenterol Jpn 1989, 24:270-276 [DOI] [PubMed] [Google Scholar]
- 30.Kanai Y, Ushijima S, Hui AM, Ochiai A, Tsuda H, Sakamoto M, Hirohashi S: The E-cadherin gene is silenced by CpG methylation in human hepatocellular carcinomas. Int J Cancer 1997, 71:355-359 [DOI] [PubMed] [Google Scholar]
- 31.Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakurs H: p16INK4 is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology 1999, 116:394-400 [DOI] [PubMed] [Google Scholar]
- 32.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S: Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis—a comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology 2000, 32:970-979 [DOI] [PubMed] [Google Scholar]
- 33.Chen YJ, Yeh SH, Chen JT, Wu CC, Hsu MT, Tsai SF, Chen PJ, Lin CH: Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology 2000, 119:431-440 [DOI] [PubMed] [Google Scholar]
- 34.Bain BJ: Human Cytogenetics: A practical Approach, ed 2, vol II. Edited by DE Rooney, BH Czepulkowski. IRL Press, 1992, pp 127–128
- 35.Shabtai F, Klar D, Kimchi D, Antebi E, Hart J, Halbrecht I: Juxta-centromeric fragility of chromosomes 1, 2, 9, 16, and immunodeficiency. Special reference to the fragility of chromosome 2 and its oncogenic potential. Anticancer Res 1984, 4:235-239 [PubMed] [Google Scholar]
- 36.Hartwell LH, Kastan MB: Cell cycle control and cancer. Science 1994, 266:1821-1828 [DOI] [PubMed] [Google Scholar]
- 37.Takai D, Yagi Y, Habib N, Sugimura T, Ushijima T: Hypomethylation of LINE1 retrotransposon in human hepatocellular carcinomas, but not in surrounding liver cirrhosis. Jpn J Clin Oncol 2000, 30:306-309 [DOI] [PubMed] [Google Scholar]
- 38.Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M: Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc Natl Acad Sci USA 1997, 94:2284-2289 [DOI] [PMC free article] [PubMed] [Google Scholar]