Abstract
The expression of thousands of genes can be monitored simultaneously using cDNA microarray technology. This technology is being used to understand the complexity of human disease. One significant technical concern regards potential alterations in gene expression because of the effect of tissue ischemia. This study evaluates the increase in the differential gene expression because of tissue processing time. To evaluate differential gene expression because of ischemia time, prostate samples were divided into five time points (0, 0.5, 1, 3, and 5 hours). Each time point consisted of a homogeneous mixture of 12 to 15 prostate tissue cubes (5 mm3). These tissues were maintained at room temperature until at the assigned time point the tissue was placed in OCT, flash frozen in liquid nitrogen, and stored at −80°C until RNA extraction. RNA from each time point was hybridized against an aliquot of 0 time point RNA from the same prostate. Four prostate glands were used in parallel studies. M-A plots were graphed to compare variability between time point sample hybridizations. Statistical Analysis of Microarray software was used to identify genes overexpressed at the 1-hour time point versus the 0-hour time with statistically significance. Microarray analysis revealed only a small percentage of genes (<0.6%) from more than 9000 to demonstrate overexpression at the 1-hour time point. Among the 41 statistically significant named overexpressed genes at the 1-hour time point were early growth response 1 (EGR1), jun B proto-oncogene (jun B), jun D proto-oncogene (jun D), and activating transcription factor 3 (ATF3). Genes previously associated with prostate cancer did not have significantly altered expression with ischemia time. Increased EGR1 protein expression was confirmed by Western blot analysis. Microarray technology has opened the possibility of evaluating the expression of a multitude of genes simultaneously, however, the interpretation of this complex data needs to be assessed circumspectly using refined statistical methods. Because RNA expression represents the tissue response to insults such as ischemia, and is also sensitive to degradation, investigators need be mindful of confounding artifacts secondary to tissue processing. All attempts should be made to process tissue rapidly to ensure that the microarray gene profile accurately represents the state of the cells and confirmatory studies should be performed using alternative methods (eg, Northern blot analysis, Western blot, immunohistochemistry).
Genome-wide scans of cancer samples has been facilitated by the use of cDNA microarray technology. This technology provides a unique method of evaluating the differential gene expression of several thousand genes simultaneously. 1,2 In cancer molecular biology this technique has been used not only to identify genes involved in tumorigenesis, but also to identify genes that may be clinically useful molecular markers. Several groups including our own have recently applied cDNA microarray technology as a tool to study prostate cancer with the goal of identifying clinically significant markers of biologically aggressive disease. 3-7 Because the technique of cDNA microarray analysis generates complex data sets, the conclusions derived are greatly dependent on the methods of data interpretation. In addition, technical concerns arise as well particularly with respect to the processing of tissue used for such experiments.
One specific area of concern is the effect of ischemia on the profile of differential gene expression. Microarray technology has identified some putative prostate cancer biomarkers such as hepsin and α-methylacyl-CoA racemase (AMACR). 3,4,8,9 However other putative candidates need to be further characterized. In this process it is important to exclude spuriously deregulated genes because of such technical artifacts such as prolonged warm ischemia before processing. This may be a more important issue within the early time period after surgical extirpation when the cellular metabolic machinery can mount a survival or apoptotic response before all metabolic activity ceases. Conversely genes that appear down-regulated could represent an artifact of RNA degradation with prolonged warm ischemia time. Some genes may also have greater susceptibility to degradation. Negative expression, therefore, is more difficult to ascertain because there is no way to distinguish decreased expression from degradation. Thus we proposed to study the increase in the differential gene expression profile throughout time of prostate tissue obtained from radical prostatectomy specimens removed as treatment for localized prostate cancer and identify individual genes that may be artifacts of processing.
Materials and Methods
With Institutional Review Board approval, four radical prostatectomy specimens were collected at the time of surgical removal and transported within 5 minutes to the surgical pathology laboratory for immediate processing. Time courses from each prostate specimen were performed independently and treated as individual experiments. Excess tissue from the clinical pathology sample was fractionated into 0-, 0.5-, 1-, 3-, and 5-hour time points. Each time point consisted of a mixture of 12 to 15 prostate tissue cubes (5 mm3) from throughout the entire prostate specimen. This was done to assure that the samples contained a homogenous mixture of stroma and epithelial cells, and benign and cancerous cells from the same prostate. These tissues were maintained at room temperature until the assigned time point and the tissue was placed in OCT, flash frozen in liquid nitrogen, and stored at −80°C until RNA extraction. In Trizol reagent (Life Technologies, Inc., Rockville, MD) tissue from the five time points were separately homogenized. RNA was obtained using a standard phenol/chloroform extraction. The integrity of total RNA was confirmed by denaturing formaldehyde agarose gel electrophoresis for each of the time points. The RNA was evaluated by spectrophotometry, qualitatively by the A260/280 ratio, and quantitatively by the absorbance at 280 nm.
The spotted glass cDNA microarray slides used in this study included ∼5000 known, named genes from the Research Genetics human cDNA clone set, 4400 expressed sequence tags. 3 Fluorescently labeled (Cy5) cDNA was prepared from total RNA from each of the ischemia time points. The reference zero (0 hour) samples used in this study were labeled using a second distinguishable fluorescent dye (Cy3) using a previously established protocol (www.microarrays.org). After labeling, the cDNA samples were neutralized, washed, and then applied to the microarray chips. After remaining in a hybridization water bath at 65°C overnight, the microarray slides were processed and scanned with a Genepix 4000 scanner.
Primary analysis was done using the Genepix software package. Images of scanned microarrays were gridded and linked to a gene print list. Initially, data were viewed as a scatter plot of Cy3 versus Cy5 intensities. Cy3 to Cy5 ratios are determined for the individual genes along with various other quality-control parameters (eg, intensity over local background). The Genepix software analysis package flags spots as absent based on spot characteristics. Furthermore, bad spots or areas of the array with obvious defects were manually flagged. Spots with small diameters (<50 μm) and spots with low signal strengths <350 fluorescence intensity units over local background in the more intense channel were discarded. Flagged spots were not included in subsequent analyses. Data are the ratio of the fluorescent cDNA probe signal hybridized against each time point to the reference 0-hour time point including the 0-hour time point itself.
Analysis introduced by ischemia time used a method, previously described, in comparing microarray quality that we have used to compare experimental samples. 10,11 Briefly, we created a M-A plot where M = log (Cy5/Cy3) is the log ratio of the two dyes used in the hybridization, and A = [log (Cy5) + log (Cy3)]/2 is the average of the log intensities. In our experiments, the Cy5-labeled time point and the Cy3-labeled 0-time reference were hybridized together. The M-A plot of the 1-hour point hybridization (1-hour Cy5 labeled versus 0-hour Cy3 labeled) is compared to the 0-time point hybridization (Cy5 labeled 0 hour versus 0-hour Cy3 labeled). We summarized the intensity data by taking the averaging Cy3 and Cy5 intensities for all of the genes for the four time courses at the 0-hour and 1-hour time points separately.
To identify statistically significant genes induced by ischemia, we analyzed our data from the four time courses with Statistical Analysis of Microarray (SAM) software (Stanford University, Stanford, CA), as previously described. 12 Briefly, SAM performs repetitive permutations based on gene-specific t-tests to identify statistically significant genes. Adjustment of a delta-tuning parameter can alter the number of false-positives, and use of threshold parameters instructs that magnitude of expression change meets cutoff requirements. Only overexpressed genes were analyzed because of the potential confounding effect of RNA degradation contributing to the number of underexpressed genes. In this experiment we used a delta of 0.20, upper threshold of 2.5-fold overexpression, and 0.4-fold under expression using a one class comparison at the 1-hour time point, because the reference in each hybridization was the 0-hour time point. The 1-hour time point was chosen because most surgical specimens are realistically processed within this time frame. Some of the genes of interest identified by SAM along with genes involved in prostate cancer were plotted together as a heat map using the Cluster/TreeView program (Stanford University).
Western blot analysis of time course was performed for one of the genes of interest, early growth response 1 (EGR1), with a readily available commercial antibody. The Western blot was performed as a time course from a single one of the prostates used in the cDNA expression microarray at the same time points. Tissues were homogenized in Nonidet P-40 lysis buffer containing 50 mmol/L Tris-HCl, pH 7.4, 1% Nonidet P-40 (Sigma, St. Louis, MO), and complete proteinase inhibitor cocktail tablet (Roche, Indianapolis, IN). After quantification using a standard Bradford assay, 15 μg of tissue protein extracts were mixed with sodium dodecyl sulfate sample buffer and electrophoresed onto a 10% sodium dodecyl sulfate-polyacrylamide gel under reducing conditions. The separated proteins were transferred onto a nitrocellulose membrane (Amersham Pharmacia Biotech, Piscataway, NJ). The membrane was incubated for 1 hour in blocking buffer [Tris-buffered saline with 0.1% Tween (TBS-T) with 5% nonfat dry milk]. The EGR1 antibody (Santa Cruz Biotechnologies, Santa Cruz, CA) was applied at 1:1000 dilution in blocking buffer overnight at 4°C. After washing three times with TBS-T buffer, the membrane was incubated with horseradish peroxidase-linked donkey anti-rabbit IgG antibody (Amersham Pharmacia Biotech) at 1:5000 for 1 hour at room temperature. The signals were visualized with the chemiluminescence enhanced chemiluminescence detection system (Amersham Pharmacia Biotech) and autoradiography. For β-tubulin control, the EGR1 antibody probed membrane was stripped with Western Re-Probe buffer (Geno-tech, St. Louis, MO) and blocked in TBS-T with 5% nonfat dry milk and incubated with rabbit anti-β-tubulin antibodies (Santa Cruz Biotechnologies) at 1:500 for 2 hours. Visualization was performed after application of the secondary antibody as described above.
Results
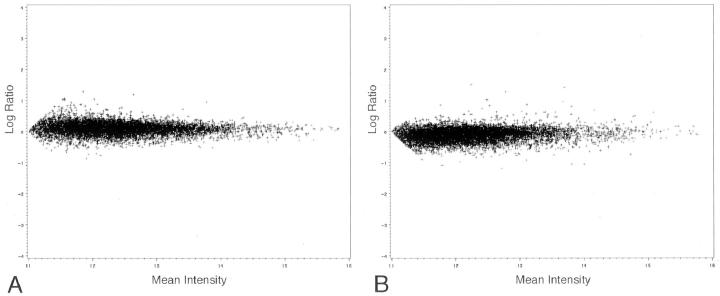
Hybridization at five time points (0, 0.5, 1, 3, and 5 hours) yielded results for each of the four prostate time courses. M-A plots from the 0- and 1-hour time points visually depict little overall variation between samples (Figure 1) ▶ . The SAM analysis identified 61 statistically significant genes that were overexpressed at the 1-hour time point. This represents only a small percentage of genes (<0.6%) with overexpression at the 1-hour time point. The 41 significant named genes are presented in Table 1 ▶ . At least several of these genes are known to be early response genes, genes implicated in hypoxia, or transcription factors, including EGR1, 13 jun B proto-oncogene (JUNB), 14 jun D proto-oncogene (JUND), 15 and activating transcription factor 3 (ATF3). 16 The expression of these genes for the four time courses throughout five time points are compared in a heat map to genes implicated in prostate cancer, such as hepsin, 3 AMACR, 8,9 fatty acid synthase, PTEN, and PIM-1 3 (Figure 2) ▶ . The gene expression for this group of cancer-related genes remains relatively constant throughout the four time courses as demonstrated by their minimal degree of red color change through the four time courses.
Figure 1.
M-A plots at the 0- and 1-hour time points. M-A plot where M = log (Cy5/Cy3) is the log ratio of the two dyes used in the hybridization, and A = [log (Cy5)+log (Cy3)]/2 is the average of the log intensities. A: The Cy5-labeled time point and the Cy3-labeled 0-time reference were hybridized together. B: The M-A plot of the 1-hour point hybridization (1-hour Cy5 labeled versus 0-hour Cy3 labeled) is compared to the 0 time point hybridization (Cy5 labeled 0 hour versus 0-hour Cy3 labeled). The data are summarized by taking the average Cy3 and Cy5 intensities for all of the genes for the four time courses at the 0-hour and 1-hour time points separately.
Table 1.
Statistically Significant Named Genes with Overexpression at 1-Hour Time Point
| Gene name | Gene ID |
|---|---|
| ZFP36, zinc finger protein 36, C3H type, homolog (mouse) | 23804 |
| CRYAB, crystalline, alpha B | 839763 |
| TTC1, tetratricopeptide repeat domain 1 | 725274 |
| EGR1, early growth response 1 | 840944 |
| JUNB, jun B proto-oncogene | 309864 |
| MT1L-OR- MT1X, metallothionein 1L-OR- metallothionein 1X | 297392 |
| USP14, ubiquitin specific protease 14 (tRNA-guanine transglycosylase) | 81599 |
| ZNF76, zinc finger protein 76 (expressed in testis) | 745003 |
| MYL2, myosin, light polypeptide 2, regulatory, cardiac, slow | 300051 |
| EGR1-OR- DCT, early growth response 1-OR- dopachrome tautomerase | 753104 |
| AGXT, alanine-glyoxylate aminotransferase | 247117 |
| JUND, jun D proto-oncogene | 767784 |
| ACADVL, acyl-coenzyme A dehydrogenase, very long chain | 810358 |
| BTG2, BTG family, member 2 | 213136 |
| SPOCK, sparc/osteonectin, cwcv and kazal-like domains proteoglycan | 754358 |
| BLVRB, biliverdin reductase B | 246035 |
| HNRPA1, heterogeneous nuclear ribonucleoprotein A1 | 511586 |
| DDM36, DDM36 | 284101 |
| ETV5, ets variant gene 5 (ets-related molecule) | 796542 |
| H2BFQ, H2B histone family, member Q | 813149 |
| UROS, uroporphyrinogen III synthase (congenital erythropoietic porphyria) | 809454 |
| HCA127, hepatocellular carcinoma-associated antigen 127 | 358549 |
| ASAHL, N-acylsphingosine amidohydrolase (acid ceramidase)-like | 324342 |
| SLC7A6, solute carrier family 7 | 767769 |
| APOC2, apolipoprotein C-II | 809523 |
| CISH, cytokine inducible SH2-containing protein | 771058 |
| EIF2S3, eukaryotic translation initiation factor 2, subunit 3 (gamma, 52kD) | 784841 |
| MRPL12, mitochondrial ribosomal protein L12 | 810173 |
| PPAP2B, phosphatidic acid phosphatase type 2B | 85394 |
| MINK, misshapen/NIK-related kinase | 756211 |
| VTN, vitronectin | 246073 |
| MB, myoglobin | 611443 |
| ATF3, activating transcription factor 3 | 51448 |
| MGC4730, similar to RIKEN cDNA 3110001D03 gene (M. musculus) | 359269 |
| HGF, hepatocyte growth factor (hepapoietin A; scatter factor) | 41650 |
| VCAM1, vascular cell adhesion molecule 1 | 49164 |
| JUN, v-jun sarcoma virus 17 oncogene homolog (avian) | 358531 |
| NUP88, nucleoporin 88kD | 843070 |
| C21orf11, chromosome 21 open reading frame 11 | 186918 |
| PCYT2, phosphate cytidylyltransferase 2, ethanolamine | 130884 |
| RALY, RNA binding protein | 825583 |
Figure 2.
TreeView analysis of benign prostate tissue demonstrating differential gene expression at different time intervals because of warm ischemia. Overexpressed and underexpressed genes are represented in red and green, respectively. Black represents no change in expression. Color intensity corresponds to the degree of overexpression or underexpression with brighter shading indicating higher level of expression.
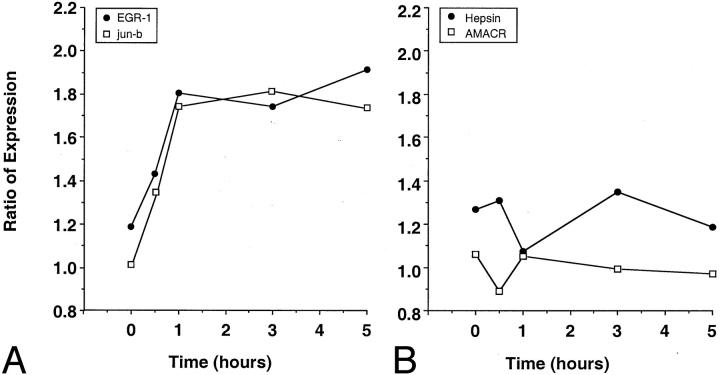
We analyzed four of the above genes in greater detail. The means of the time point expression ratios (each time point hybridized against 0 time) for all four prostate time courses was plotted for two of the genes identified as significant by the SAM analysis, EGR1 and jun B (Figure 3A) ▶ . The gene expression throughout time for these two genes increases to a steady level by 3 hours. Whereas, the expression of two genes implicated as prostate cancer markers, hepsin and AMACR, remains steady throughout the time course (Figure 3B) ▶ .
Figure 3.
cDNA microarray gene expression intensity of select genes throughout time course. A: Genes such as EGR-1 and jun-b demonstrated near twofold increase throughout time because of ischemia. B: Other genes previously reported as putative prostate cancer biomarkers demonstrated little change throughout the same time course.
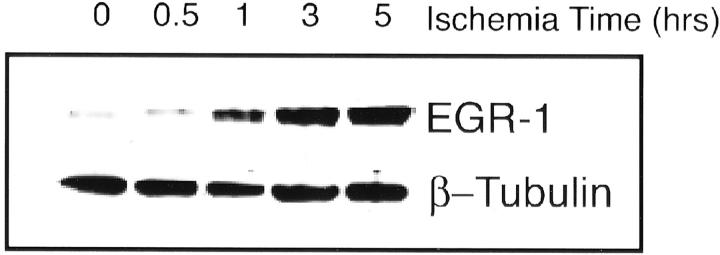
Confirmatory Western blot analysis with an anti-EGR1 antibody is presented in Figure 4 ▶ , where each lane corresponds to a point in the time course. A band at 58 kd corresponds to the expected weight of the 543-amino acid-long EGR1 protein. The intensity of the bands increases throughout the time course, implying increased EGR1 protein expression with time.
Figure 4.
Western blot analysis with anti-EGR1 antibody throughout the time course (0, 0.5, 1, 3, and 5 hours) confirms increased EGR-1 protein accumulation throughout time, confirming the expression array data. β-tublin serves as a control for protein loading.
Discussion
cDNA microarray technology has become a widely used tool for molecular profiling. The techniques used to analyze the vast quantity of data generated are variable, but systematic statistical methods for analyzing the data and accounting for the inherent variability of the experiment (microarray chip quality and differential labeling) are being introduced. Little research thus far has been devoted to systemic variability contributed by the samples themselves beyond the heterogeneity involved in using biological tissue.
To our knowledge, Huang and colleagues 17 were the first to study the effects of ischemia on the gene expression profile probed by cDNA microarrays. These investigators were interested in the general pattern of expression in colon cancer changes secondary to the ischemic effect. They used a latent class model to observe a change from the average gene expression. This study preliminarily established that ischemia alters the gene expression profile in tissue, but was severely limited by analyzing only one specimen and using a cell line as the reference.
Our study is the first to introduce the concept of systemic sample effect. In our experiments this effect was because of the ischemia time of tissue processing. By performing our time-course experiments independently on four samples, we have mitigated the problem of false-positives introduced by a lack of replicates to identify a group of genes that are overexpressed in prostate tissue solely because of ischemia. 18 For at least one of these genes, EGR1, we were also able to demonstrate increased protein expression throughout time.
Our study presumed that there would be altered gene expression throughout time; interestingly however, there was little overall increase in the gene expression variability present with ischemia time, as depicted in the M-A plots. We assumed that we would not be able to distinguish degradation from decreased expression; therefore we pursued genes with a pattern of expression that peaked at 1 hour. In our radical prostatectomy harvesting protocol, most surgical specimens are routinely processed within 30 minutes after surgical extirpation. However, keeping in mind the logistics of transporting the freshly extirpated specimen, and accessioning, examining, and sectioning the specimen, we believed that 1 hour would be a realistic time frame before the specimen can be frozen to prevent further RNA degradation. With this time course in mind, we identified four reference genes: EGR1, Jun B, Jun D, and ATF3 that increased in expression throughout time. The longer time points were examined as extreme examples for radical prostatectomy samples. However, in some of our prostate cancer-profiling work, we have used advanced tumor from a rapid autopsy program, in which samples may be harvested up to 3 to 5 hours after death. 19
Several studies have now begun to exploit the high-throughput capabilities of cDNA microarray to obtain a molecular profile of prostate cancer. 3-7,20 Several interesting genes that play a putative role in prostate cancer have been identified. These notably include two genes that have aroused interest recently, hepsin 3-6 and AMACR. 8,9 The usefulness of the present study is to evaluate whether the processing time may influence the gene expression profile for prostate tissue specimens. Although we identified several genes with a statistically significant increase in expression at 1 hour, none of the recently reported genes involved in prostate cancer development appeared to be dramatically affected by ischemia time.
Of the 41 significant named genes that met our criteria for overexpression at 1 hour, several have been identified in the literature for increased expression secondary to ischemic stress. Jun B and jun D transcript levels are elevated after cerebral ischemia in animal models. 14,15 Activating transcription factor 3 (ATF3) is induced by ischemia in a variety of tissues including heart, liver, kidney, and brain. 16 In most tissue types ATF3 mRNA expression is increased within 2 hours of the insult. 16 EGR1 appears to function as a master switch to activate several cellular responses to ischemic stress rapidly after the insult. 13 Most of the overexpressed genes we identified were not related to prostate cancer; except for one of these genes, EGR1. 21,22 Eid and colleagues 21 compared EGR1 expression in prostate cancer from transurethral prostatectomy and total prostatectomy specimens from men with untreated disease versus normal prostate obtained during autopsy of organ donors. EGR1 mRNA expression was elevated in prostate cancer by Northern blot analysis. In situ hybridization confirmed the finding and showed expression mainly confined to the epithelium. Quantitative in situ hybridization revealed increased EGR1 expression with higher Gleason grade cancer. Finally EGR1 protein was confined to the epithelium by immunohistochemistry, a finding confirmed by others. 22 In our study, in which the relative expression of all genes were compared to their 0-hour time point for all time points, we found EGR1 to be exemplary of a gene with increased expression with ischemia time. In contrast, the relative expression of hepsin, a gene whose overexpression has been confirmed in several studies and been validated by Northern blot analysis and immunohistochemistry, 3 remained constant throughout the time course. That two genes previously identified with prostate cancer could have different expression patterns with ischemia time would imply that some prostate cancer genes might be more susceptible to ischemia. EGR1 in particular may be such a gene because of its central role in the cellular response to ischemia. 13 Alternatively, increased gene expression is the artifact of tissue processing that could have begun with tissue ischemia while ligating the blood supply to the prostate during surgery that continued until freezing, although this source of error should have been minimized by using a 0-time point as the reference during hybridizations.
Based on the work of Huang and colleagues 23 and the current study, confirmation of differentially expressed genes by cDNA analysis is critical. In our study a pattern of gene expression including EGR1, jun B, jun D, and ATF 3 suggests an ischemia effect. We recommend analyzing prostate cancer cDNA microarray data circumspectly if this pattern is present. Furthermore, in this study we validated EGR1 expression with Western blot analysis, but various other confirmation techniques are available and should be used before overstating the potential role of a biomarker.
Conclusion
Microarray technology is a tool that has opened the possibility of evaluating the expression of a multitude of genes simultaneously, however, the interpretation of this complex data needs to be assessed circumspectly using refined statistical methods. Because RNA expression represents the tissue response to insults such as ischemia, and is also sensitive to degradation, we further caution investigators to be mindful of confounding artifacts secondary to tissue processing. All attempts should be made to process tissue rapidly to ensure that the microarray gene profile accurately represents the state of the cells. Confirmation should also be sought using alternative methods (eg, Northern blot analysis, Western blot, immunohistochemistry).
Footnotes
Address reprint requests to Mark A. Rubin, M.D., Brigham and Women’s Hospital, Pathology, 75 Francis Street, Boston, MA 02115. E-mail: marubin@partners.org.
Supported by the Specialized Program in Research Excellence in Prostate Cancer (P50 CA69568) and the National Cancer Institute (to M. A. R. and A. M. C.).
A. T. and I. P. M. contributed equally to this work.
References
- 1.Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM: Expression profiling using cDNA microarrays. Nat Genet 1999, 21:10-14 [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO: Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270:467-470 [DOI] [PubMed] [Google Scholar]
- 3.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM: Delineation of prognostic biomarkers in prostate cancer. Nature 2001, 412:822-826 [DOI] [PubMed] [Google Scholar]
- 4.Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, Catalona WJ, Watson MA, Milbrandt J: Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res 2001, 61:5692-5696 [PubMed] [Google Scholar]
- 5.Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB: Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res 2001, 61:4683-4688 [PubMed] [Google Scholar]
- 6.Stamey TA, Warrington JA, Caldwell MC, Chen Z, Fan Z, Mahadevappa M, McNeal JE, Nolley R, Zhang Z: Molecular genetic profiling of Gleason grade 4/5 prostate cancers compared to benign prostatic hyperplasia. J Urol 2001, 166:2171-2177 [PubMed] [Google Scholar]
- 7.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson Jr HF, Hampton GM: Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res 2001, 61:5974-5978 [PubMed] [Google Scholar]
- 8.Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM: Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res 2002, 62:2220-2226 [PubMed] [Google Scholar]
- 9.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM: Alpha-methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA 2002, 287:1662-1670 [DOI] [PubMed] [Google Scholar]
- 10.Dudoit S, Yang YH, Callow MJ, Speed TP: Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Technical Report 578. 2000:p 38 UC Berkeley, Berkeley CA
- 11.Tseng GC, Oh MK, Rohlin L, Liao JC, Wong WH: Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res 2001, 29:2549-2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001, 98:5116-5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan SF, Fujita T, Lu J, Okada K, Shan Zou Y, Mackman N, Pinsky DJ, Stern DM: Egr-1, a master switch coordinating upregulation of divergent gene families underlying ischemic stress. Nat Med 2000, 6:1355-1361 [DOI] [PubMed] [Google Scholar]
- 14.Hara T, Mies G, Hata R, Hossmann KA: Gene expressions after thrombolytic treatment of middle cerebral artery clot embolism in mice. Stroke 2001, 32:1912-1919 [DOI] [PubMed] [Google Scholar]
- 15.Kamme F, Wieloch T: Induction of junD mRNA after transient forebrain ischemia in the rat. Effect of hypothermia. Brain Res Mol Brain Res 1996, 43:51-56 [DOI] [PubMed] [Google Scholar]
- 16.Hai T, Hartman MG: The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 2001, 273:1-11 [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Choudhri TF, Winfree CJ, McTaggart RA, Kiss S, Mocco J, Kim LJ, Protopsaltis TS, Zhang Y, Pinsky DJ, Connolly Jr ES: Postischemic cerebrovascular E-selectin expression mediates tissue injury in murine stroke. Stroke 2000, 31:3047-3053 [PubMed] [Google Scholar]
- 18.Lee ML, Kuo FC, Whitmore GA, Sklar J: Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc Natl Acad Sci USA 2000, 97:9834-9839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ: Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res 2000, 6:1038-1045 [PubMed] [Google Scholar]
- 20. Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR: Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002, 1:203-209 [DOI] [PubMed] [Google Scholar]
- 21.Eid MA, Kumar MV, Iczkowski KA, Bostwick DG, Tindall DJ: Expression of early growth response genes in human prostate cancer. Cancer Res 1998, 58:2461-2468 [PubMed] [Google Scholar]
- 22.Abdulkadir SA, Carbone JM, Naughton CK, Humphrey PA, Catalona WJ, Milbrandt J: Frequent and early loss of the EGR1 corepressor NAB2 in human prostate carcinoma. Hum Pathol 2001, 32:935-939 [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Qi R, Quackenbush J, Dauway E, Lazaridis E, Yeatman T: Effects of ischemia on gene expression. J Surg Res 2001, 99:222-227 [DOI] [PubMed] [Google Scholar]