Abstract
In bacterial and phage genomes, coding regions are sometimes interrupted by self-splicing introns or inteins, which can encode mobility-promoting homing endonucleases. Homing endonuclease genes are also found free-standing (not intron- or intein-encoded) in phage genomes where they are inserted in intergenic regions. One example is the HNH family endonuclease, mobE, inserted between the large (nrdA) and small (nrdB) subunit genes of aerobic ribonucleotide reductase (RNR) of T-even phages T4, RB2, RB3, RB15, and LZ7. Here, we describe an insertion of mobE into the nrdA gene of Aeromonas hydrophila phage Aeh1. The insertion creates a unique genes-in-pieces arrangement, where nrdA is split into two independent genes, nrdA-a and nrdA-b, each encoding cysteine residues that correspond to the active-site residues of uninterrupted NrdA proteins. Remarkably, the mobE insertion does not inactivate NrdA function, although the insertion is not a self-splicing intron or intein. We copurified the NrdA-a, NrdA-b, and NrdB proteins as complex from Aeh1-infected cells and also showed that a reconstituted complex has RNR activity. Class I RNR activity in phage Aeh1 is thus assembled from separate proteins that interact to form a composite active site, demonstrating that the mobE insertion is phenotypically neutral in that its presence as an intervening sequence does not disrupt the function of the surrounding gene.
Keywords: bacteriophage Aeh1, gene structure, intervening sequence
Homing endonucleases are a distinctive class of site-specific DNA endonucleases that promote the lateral transfer of their own coding region and flanking DNA between genomes by a recombination-dependent process termed homing (reviewed in ref. 1). Homing endonucleases are often encoded within self-splicing introns and inteins (2–4), but many bacterial and phage genomes possess a significant number of so-called free-standing homing endonucleases that are not encoded within introns or inteins (5, 6). Free-standing endonucleases do not have the benefit of a self-splicing element to minimize their impact on host gene structure and function and, thus, are found at genomic insertion sites that are of low impact, such as intergenic regions. In T4-like phages that infect Escherichia coli and related bacteria, free-standing endonucleases are more abundant than intron-encoded versions (6–9) and promote their spread to phage genomes lacking the endonuclease by an intronless homing pathway (10–12).
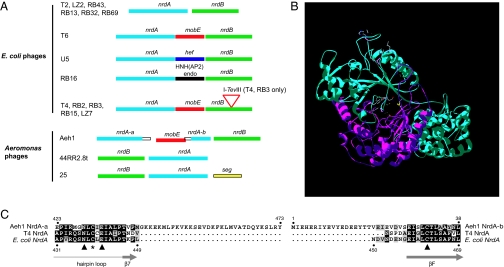
As a consequence of their abundance in T4-like phages, free-standing endonucleases represent a significant source of genetic variation by promoting recombination between genomes. Many characterized homing endonucleases have recognition sites that lie in genes that function in DNA metabolism, including those that encode aerobic and anaerobic ribonucleotide reductases (RNRs) (13). Three classes of RNRs have been described to date, based on required metallocofactors used to generate a radical intermediate and on structural differences (14). Prokaryotic class Ia aerobic enzymes, typified by the Escherichia coli nrdA and nrdB genes, are α2β2 heterodimers of the large NrdA (α) and small NrdB (β) proteins, respectively (15). Moreover, the nrdA and nrdB genes are well conserved, offering a good target for homing endonucleases. Consistent with this prediction, many phage- and bacterial-encoded RNR genes are interrupted by self-splicing introns or inteins, with the insertion sites often near the active site of the enzymes (16–18). In phages T4 and RB3, the nrdB gene is interrupted by a group I intron encoding a HNH family homing endonuclease, I-TevIII (19–21). Of relevance to this study is the free-standing HNH endonuclease, mobE, that is inserted between the nrdA and nrdB genes of phages T4, RB2, RB3, RB15, and LZ7 (9, 22) (Fig. 1A). MobE efficiently spreads between T-even phages, and likely possesses a cleavage site within or nearby nrdA or nrdB (12, 22). The nrdA-nrdB region of T-even phages thus appears to be a hotspot of homing endonuclease-mediated recombination.
Fig. 1.
The nrdA-nrdB genomic region of T-even phage is a hotspot of homing endonuclease insertion. (A) Gene composition of the nrdA-nrdB region of E. coli and Aeromonas phages (not to scale). Self-splicing group I introns are indicated by a triangle. Open boxes indicate the unique DNA sequences accompanying the mobE insertion. HNH(AP2), HNH family endonuclease (47); hef, homing endonuclease-like function (22); seg, similarity to endonuclease encoded within group I introns (5). (B) Homology model of Aeh1 NrdA-a and NrdA-b proteins based on the known structure of E. coli NrdA (24) by using the program Swiss-Model at ExPASy Home Page (http://swissmodel.expasy.org/). The RNR-specific β/α-barrel domain with the active site is a composite of the NrdA-a (blue) and NrdA-b (magenta). Active-site residues Cys-219, Asn-429, Cys-431, and Glu-433 in NrdA-a and Cys-31, Tyr-300, and Tyr-301 in NrdA-b (model center) are highlighted as well as conserved specificity-site residues Asps226, Leu-228, Ile-262, His-269, and Phe-275 in NrdA-a (model top). (C) Amino acid alignment of the split Aeh1 NrdA-a and NrdA-b proteins with the contiguous E. coli and phage T4 NrdA homologs. Numbers above the alignment indicate amino acid positions in the Aeh1 NrdA-a and NrdA-b proteins, whereas numbers below the alignment refer to the E. coli NrdA protein. The star indicates a conserved active-site cysteine residue (Cys-439 of E. coli NrdA), and triangles indicate conserved active-site residues (from left Asn-437, Glu-441, and Cys-462 of E. coli NrdA). Secondary-structure elements of E. coli NrdA are indicated below the alignment (β, β-strand). Black and gray shading indicate identical and similar amino acids, respectively, between the sequences.
Here, we examine the impact of free-standing endonucleases on gene structure and function. We describe a gene arrangement created by the insertion of mobE into the coding region of the nrdA gene of Aeromonas hydrophila T-even phage Aeh1 that possesses the characteristics of a transposition event. The mobE insertion fragments the nrdA gene at the active site, creating two independent genes that each encode key active site residues of NrdA proteins. Remarkably, the mobE insertion is not inactivating for RNR function, and we show that the split NrdA-a and NrdA-b proteins assemble into a complex with the NrdB protein that retains RNR activity. Our results demonstrate that the mobE insertion is phenotypically neutral with respect to NrdA function and that, despite the absence of splicing, the presence of an intervening sequence does not disrupt the function of the surrounding gene.
Results
A Split nrdA Gene in Phage Aeh1 Created by the Insertion of an HNH Homing Endonuclease.
Database searches revealed a mobE homolog in the sequenced genome of the A. hydrophila T4-like phage Aeh1 (6, 23). Intriguingly, the Aeh1 mobE gene is inserted in a different genomic position relative to mobE endonucleases in phages T4, RB2, RB3, RB15, and LZ7, where it is inserted between nrdA and nrdB (Fig. 1A). The Aeh1 mobE insertion interrupts the nrdA gene, splitting it into two smaller coding regions, nrdA-a and nrdA-b that are in different reading frames relative to each other. Associated with the insertion is ≈100 bp of unique DNA fused in-frame to the 3′ end of the nrdA-a gene and ≈40 bp of unique DNA fused in-frame to the 5′ end of the nrdA-b gene [see supporting information (SI) Fig. 5]. This unique sequence is not present in characterized nrdA genes, does not flank the mobE gene in phages T4, RB2, RB3, RB15, and LZ7, and does not match any sequence in public databases as judged by BLAST searches. The Aeh1 mobE insertion is unique, because, in other organisms, the nrdA gene is a contiguous coding region (currently >400 sequences in GenBank, available at the Ribonucleotide Reductase Database, http://rnrdb.molbio.su.se).
We built a homology model of the split Aeh1 NrdA-a and NrdA-b proteins using the E. coli NrdA structure as a reference (24) (Fig. 1B). The mobE insertion splits the Aeh1 nrdA gene between two adjacent β-strands in the RNR-specific β/α-barrel that constitutes the active site (Fig. 1B). Significantly, the two conserved active-site cysteine residues, Cys-219 in NrdA-a and Cys-31 in NrdA-b, are located in separate genes (Fig. 1 B and C). The homologous residues in E. coli NrdA (Cys-225 and Cys-462) are within ≈6 Å (24). The mobE insertion also results in the separation of two tyrosines (Tyr-300 and Tyr-301 of NrdA-b) from a third active site cysteine (Cys-431 in NrdA-a). Previous work has demonstrated that these conserved tyrosines play a critical role in the transfer of the radical from NrdB to the catalytic cysteines in NrdA (25). A functional class Ia RNR in phage Aeh1 must therefore involve assembly of the β/α-barrel from the split large subunit gene products bringing the active site residues in close proximity to form a composite active site and interact with the small NrdB subunit to form a functional holoenzyme complex. Because the mobE insertion could have obvious implications for class I RNR activity in phage Aeh1, we focus here on determining whether the insertion is compatible with RNR function.
The mobE Insertion Is Not a Self-Splicing Intron or Intein.
In considering how a functional RNR activity might arise, we noted that endonuclease-containing self-splicing introns and inteins are common in bacterial and phage RNR genes (16–18, 21), suggesting that RNA- or protein-splicing could assemble a contiguous nrdA gene. We could not, however, identify sequence elements typical of self-splicing introns or inteins at the putative splice junctions (4, 26). Nevertheless, it is possible that the mobE insertion represents a divergent or previously unidentified type of intron or intein (27–29).
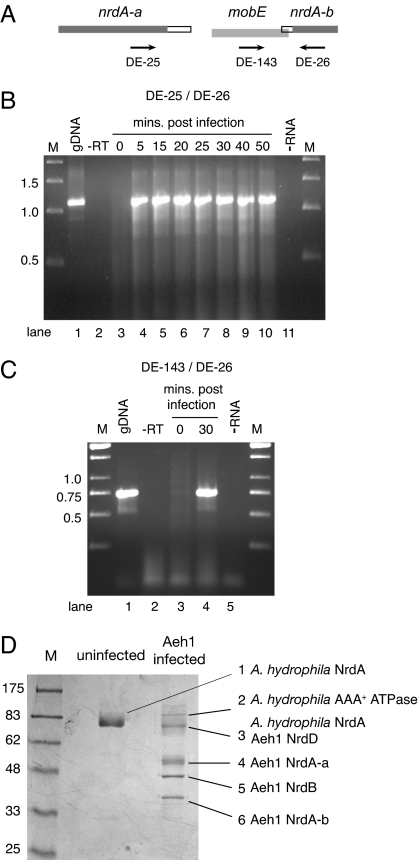
To determine whether RNA splicing removed the mobE insertion, we performed RT-PCR on total RNA isolated before and after Aeh1 infection. With primers flanking the mobE insertion (Fig. 2A), a 1.1-kb RT-PCR product was amplified at all time points sampled after phage infection (Fig. 2B, lanes 4–10). This product was the same size as that amplified from phage genomic DNA (Fig. 2B, lane 1). We sequenced the RT-PCR product and confirmed that the sequence was identical to the genomic sequence. A second RT-PCR was performed such that a product would be amplified only when the mobE insertion was not removed by RNA splicing (Fig. 2C). Using RNA isolated 30-min after infection, we amplified an RT-PCR product of 0.73 kb (Fig. 2C, lane 4), the same size as the genomic amplification product (Fig. 2C, lane 1). A similar result was obtained using RNA isolated at earlier time points after Aeh1-infection (not shown). Thus, two independent RT-PCRs demonstrate that the mobE insertion is not a self-splicing intron and that the nrdA-a, mobE and nrdA-b genes are cotranscribed.
Fig. 2.
The Aeh1 mobE insertion is not a self-splicing intron or intein. (A) Schematic of the Aeh1 mobE insertion indicating the approximate position of primers. (B) RT-PCR with primers DE-25/DE-26. Shown is a 1% agarose gel of aliquots of RT-PCRs using total RNA isolated at the indicated times after Aeh1 infection. gDNA, PCR with Aeh1 genomic DNA; −RT, PCR performed without prior reverse transcriptase step; −RNA, reaction performed without RNA; M, molecular weight markers with sizes indicated in kilobases. (C) RT-PCR with primers DE-143/DE-26 and labeled as in B. (D) Purification of phage Aeh1-encoded RNR proteins from infected A. hydrophila cell extracts by dATP-Sepharose chromatography. Shown is a Coomassie-stained 10% SDS/PAGE gel of a dATP-Sepharose purification using uninfected or Aeh1-infected A. hydrophila cell extracts. Bands corresponding to proteins identified by mass spectrometry are numbered. M, molecular mass marker with sizes indicated in kilodaltons.
To determine whether protein splicing created a contiguous NrdA protein, we used dATP-Sepharose chromatography to purify the protein from phage-infected A. hydrophila cell extracts (30–32). Class I aerobic RNRs are allosterically regulated by dATP, which binds to a site in the NrdA subunit (24). Based on sequence similarity to the E. coli enzyme, the dATP-binding site is located in the Aeh1 NrdA-a protein. As shown in Fig. 2D, five polypeptides eluted from the dATP-Sepharose column when an Aeh1-infected cell extract was applied (Fig. 2D, Aeh1 infected). One of these polypeptides (band 4) was positively identified as the Aeh1 NrdA-a protein by mass spectrometry, consistent with this polypeptide possessing a dATP-binding site. Significantly, two additional polypeptides (bands 5 and 6) were identified by mass spectrometry as the Aeh1-encoded NrdB and NrdA-b proteins, respectively. As expected, the A. hydrophila NrdA protein was also purified from both uninfected (Fig. 2D, uninfected, band 1) and Aeh1-infected cells (Fig. 2D, band 3). Mass spectrometry of band 3 revealed a mixture of the A. hydrophila NrdA protein and the Aeh1-encoded NrdD protein, the anaerobic RNR.
Purification of proteins that correspond to the predicted sizes of the Aeh1-encoded NrdA-a and NrdA-b proteins is significant because it rules out intein splicing or ribosomal hoping (33, 34) as a mechanism to restore a contiguous large subunit protein. Our data also show that the nrdA-a and nrdA-b genes have their own ribosome-binding sites (SI Fig. 5) and are independently translated from a common transcript. Moreover, the observation that the Aeh1 NrdA-a, NrdA-b and NrdB proteins coelute from the dATP-Sepharose column implies that they form a tight complex, and that formation of this complex likely occurs by a posttranslational mechanism.
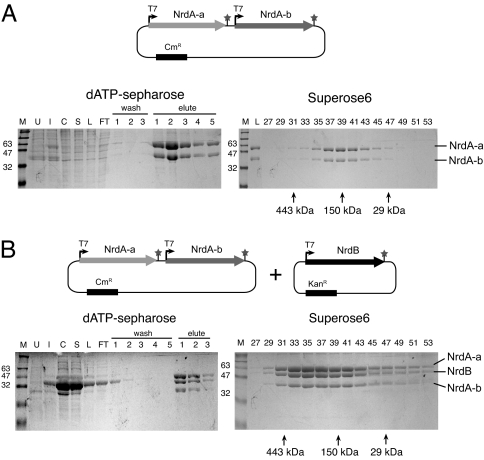
The NrdA-a and NrdA-b Proteins Assemble Independently of Interactions with NrdB.
To facilitate further characterization of the split Aeh1 RNR gene products, we cloned the Aeh1 nrdA-a, nrdA-b, and nrdB genes into compatible E. coli-based overexpression vectors. We found that the NrdA-a or NrdA-b proteins were insoluble when individually expressed, but coexpression of the two proteins increased solubility (not shown). We thus transformed an E. coli expression strain with a pNrdA-a/NrdA-b dual-expression plasmid (Fig. 3A) to ask whether the proteins could interact and form a heterodimeric complex independent of interactions with the NrdB small subunit. As shown in Fig. 3A, the NrdA-a and NrdA-b proteins coeluted from the dATP-Sepharose column. To confirm that the NrdA-a and NrdA-b proteins formed a complex, we applied the peak dATP-Sepharose fraction to a gel-filtration column. Both proteins coeluted, and we estimated the molecular mass of the NrdA-a/NrdA-b complex to be ≈186 kDa with a ≈1:1 stoichiometry of NrdA-a:NrdA-b (Fig. 3A), in agreement with a predicted 181 kDa (α-a/α-b)2 dimer of heterodimers. We repeated the above experiment with the NrdA-a and NrdA-b proteins expressed in trans from two compatible E. coli plasmids and obtained similar results (not shown). Collectively, these results show that the NrdA-a and NrdA-b proteins form a complex independent of interactions with the NrdB protein.
Fig. 3.
Reconstitution of RNR complexes using purified proteins. (A) Coexpression and purification of NrdA-a and NrdA-b by dATP-Sepharose and Superose6 gel-filtration chromatography. A schematic of the pNrdA-a/NrdA-b expression plasmid is shown, with T7 RNA polymerase promoters (T7) indicated by right-facing arrows and transcriptional termination signals indicated by stars. Shown are 12% SDS/PAGE gels with aliquots of each purification step. Fractions from wash or elution steps are indicated above the gel. M, molecular mass standards (in kilodaltons); U, uninduced extract; I, induced extract; C, crude lysate; S, soluble fraction; L, column load; FT, column flowthrough. (B) Coexpression and purification of NrdA-a, NrdB, and NrdA-b, labeled as in A.
Reconstitution of an Active RNR Complex.
We next coexpressed the NrdA-a, NrdA-b, and NrdB proteins in the same E. coli strain (Fig. 3B). All three proteins coeluted from the dATP-Sepharose column, consistent with the purification of the three proteins from Aeh1-infected cells (Fig. 2D). We separately purified the NrdB protein by ion-exchange chromatography, and determined that the purified protein did not bind to the dATP-Sepharose column (data not shown), indicating that coelution of NrdB with NrdA-a and NrdA-b from the dATP-Sepharose column was due to its interactions with the large subunit proteins. The dATP-Sepharose fractions were applied to a gel-filtration column, and we found that all three proteins coeluted in the same fractions, independent of the initial concentration of the sample (Fig. 3B). Estimation of the size of the NrdA–a/NrdA–b/NrdB complex was problematic, however, because the complex eluted over a broad range of fractions, and because the elution profile of the complex depended on the initial sample concentration. At 0.1 mg/ml, the size of the complex was estimated to be ≈256 kDa, with the NrdB and NrdA-b proteins present in a 0.89:0.95 ratio relative to NrdA-a, suggesting a (α-a/α-b) 2β2 structure mimicking the α2β2 bacterial class I RNRs. The separately purified NrdB protein was also applied to the gel-filtration column, and found to elute in a different range of fractions than those of the NrdA-a/NrdA-b complex, and the NrdA-a/NrdA-b/NrdB complex (data not shown).
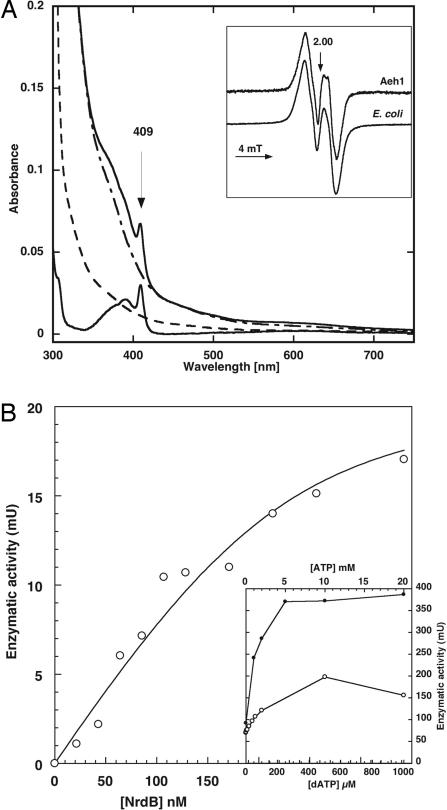
We assayed the purified proteins for aerobic RNR activity. No RNR activity was detected with the NrdA-a/NrdA-b complex alone, or with the NrdB protein by itself (Table 1). Instead, we observed RNR activity when the separately purified NrdA-a/NrdA-b and NrdB proteins were combined (Table 1). The purified NrdB required reconstitution by iron to acquire the typical diiron/tyrosyl radical cofactor (0.47 radical/dimer) (Fig. 4A). Using the reconstituted NrdB protein and DTT as an artificial electron donor, we observed a specific activity of close to 100 units/mg protein for the complex, and the dissociation constant for NrdA-a/NrdA-b versus NrdB was estimated to be 0.03 μM (Fig. 4B). Enzymatic activity was allosterically stimulated by both ATP (KL = 1.2 mM) and dATP (KL = 0.12 mM) (Fig. 4B Inset). The Aeh1 NrdA-a protein appears to have an N-terminal ATP-cone domain, which is responsible for dATP-dependent inhibition of enzyme activity in most RNRs (14, 24). However, two critical histidine residues responsible for this inhibition are replaced by Gln-54 and Ala-83 in the Aeh1 NrdA-a protein (15), which plausibly explains its lack of dATP-dependent inhibition. Importantly, the Aeh1 NrdA-a/NrdA-b and NrdB components showed no cross reactivity with the A. hydrophila NrdA protein, or with the E. coli NrdA or NrdB proteins (Table 1). Collectively, these results show that the NrdA-a, NrdA-b, and NrdB proteins copurify through two columns, form a tight complex, and exhibit RNR activity.
Table 1.
The Aeh1 NrdA-a/NrdA-b/NrdB complex has high enzyme activity
| Added protein | Specific enzyme activity, units/mg |
|
|---|---|---|
| Aeh1 NrdA-a/NrdA-b | Aeh1 NrdB | |
| Aeh1 NrdA-a/NrdA-b | 0.8* | 94 ± 1 |
| Aeh1 NrdB | 94 ± 1 | 0.3* |
| A. hydrophila NrdA | n.a. | 2.4 ± 0.3† |
| E. coli NrdA | n.a. | 1.3 ± 0.02 |
| E. coli NrdB | 0.5 ± 0.01 | n.a. |
Amounts of protein used were 4 μg of Aeh1 NrdA-a/NrdA-b, Aeh1 NrdB, A. hydrophila NrdA, and E. coli NrdB, and 1 μg of E. coli NrdA. n.a., not applicable.
*Background enzyme activity of 4 μg of Aeh1 NrdA-a/NrdA-b, or 4 μg of Aeh1 NrdB.
†Background activity of 4 μg of A. hydrophila NrdA was 1.8 units/mg.
Fig. 4.
Enzyme activity of Aeh1 NrdA-a/NrdA-b and NrdB proteins. (A) Uv-vis spectra of 18 μM Aeh1 NrdB protein: as purified (dashed line), after iron reconstitution (upper solid line), after iron reconstitution and scavenging of tyrosyl radical by 10 mM hydroxyurea treatment (dash-dotted line), subtraction of before and after hydroxyurea treatment (lower solid line). (Inset) Electron paramagnetic resonance spectrum of iron reconstituted Aeh1 NrdB (45 μM protein, 0.47 radical/dimer) and E. coli NrdB. (B) Titration of enzyme activity in presence of increasing concentrations of Aeh1 NrdB. Curve fitting was performed as described (42) and gave KD 0.034 ± 0.057 μM (the enzyme activity level required an Aeh1 NrdA-a/NrdA-b concentration of 0.22 μM, which results in high standard error). (Inset) Titration of enzyme activity in presence of increasing concentrations of ATP (filled circles) and dATP (open circles).
Discussion
Phage genomes are littered with free-standing homing endonuclease genes (6, 7, 9), characterized examples of which function as selfish genetic elements by promoting their spread between phage genomes (10–12). Many free-standing endonucleases are found at genomic insertion sites that are tolerated by the host because they are of low impact, such as intergenic regions. Here, however, we describe an insertion of a homing endonuclease gene, mobE, into a conserved and critical gene of DNA metabolism, the large subunit gene (nrdA) of aerobic RNR. Significantly, the nrdA gene of phage Aeh1 is split such that active site residues in the β/α-barrel domain are encoded in two separate genes, nrdA-a and nrdA-b, a unique organization for RNR genes. We show that, despite the mobE insertion, phage Aeh1 possesses a functional class I RNR enzyme with a composite active site formed by residues contributed by the NrdA-a and NrdA-b proteins in complex with the NrdB protein. Our results have implications for the evolution of structure and function of RNRs, and more generally suggest that novel catalytic sites could be created from polypeptides that have been shuffled between genomes as a consequence of homing endonuclease mobility.
The observation that T4-like phages related to Aeh1 have an uninterrupted nrdA coding region strongly suggests that the Aeh1 split gene structure resulted from the insertion of mobE into nrdA. Furthermore, the intergenic location of mobE in phages T4, RB2, RB3, RB15, and LZ7 also supports a scenario whereby mobE invaded a contiguous nrdA-coding region in a phage ancestral to Aeh1 (6, 22). The alternative hypothesis, that the nrdA gene was already split, and mobE invaded the nrdA-a and nrdA-b intergenic region is unlikely, because nrdA genes are contiguous in all organisms examined to date. Although the recognition and cleavage sites of mobE are uncharacterized, inheritance of genetic markers in phage crosses indicate that the mobE cleavage sites are likely located within or nearby the nrdA or nrdB genes (22), suggesting that a rare transposition event may have resulted in mobE invading the Aeh1 nrdA gene. Transposition of homing endonucleases to nonallelic sites is thought to result from the sequence-tolerant binding ability of homing endonucleases, facilitating binding and cleavage at sites that share nucleotide similarity with the endonuclease's primary recognition site. Illegitimate DSB-repair of the cleaved non-allelic site by using short stretches of DNA homology surrounding the endonuclease's insertion site may provide a mechanism by which the endonuclease could transpose to a new genomic location (35).
Because many characterized homing endonucleases have recognition and cleavage sites within protein-coding genes, transposition of free-standing endonucleases into coding regions by illegitimate DSB repair may occur at an appreciable rate (36–38). A Seg-like homing endonuclease is found associated with a split gene structure in the family B DNA polymerase (gp43) of Aeromonas salmonicida phage 25 (23, 39). Related phage possess a split gene 43 separated by up to ≈3 kb of intervening sequence without a Seg-like endonuclease. The insertions in gene 43 occur such that the DNA polymerases are split between two defined units of protein structure, separating amino acid residues involved in polymerase function; presumably these split gene products must assemble to form a functional enzyme. A similar situation is observed in the case of the T4-encoded type II topoisomerase, which is split into two smaller genes (39 and 60) by the insertion of an HNH endonuclease, mobA (7) (D. Shub, personal communication).
Previously characterized bacterial class I RNR enzymes are a α2β2 complex composed of NrdA (α) and NrdB (β) homodimers (15), although recent characterization of a P. aeruginosa class I enzyme suggested a higher oligomeric structure (40). In phage Aeh1, however, we show that the RNR holocomplex is composed of two split large subunit proteins, NrdA-a and NrdA-b, and the small subunit protein, NrdB. Interestingly, a documented example of a split RNR gene involves a class II RNR enzyme, nrdJ, in P. aeruginosa and other γ-proteobacteria (41). The two halves of nrdJ are translated separately, and class II RNR activity was detected in P. aeruginosa cell extracts. The split, however, is not at the active site of the enzyme (as is described here), and is not created by the insertion of a homing endonuclease.
Our data raise the intriguing question as to the nature of the mechanism(s) by which active site residues on the separate NrdA-a and NrdA-b proteins are assembled correctly to form a composite active site domain and a fully functional large subunit. This mechanism must involve protein-protein interactions, because we find no evidence of RNA or protein splicing to remove the mobE insertion from the nrdA-a/mobE/nrdA-b transcript, and because we can purify a NrdA-a/NrdA-b/NrdB complex from Aeh1-infected cells. In this regard, the dimerization interface of the E. coli NrdA subunit has been well characterized (15), and the amino acid residues involved in dimerization are conserved in the phage Aeh1 NrdA-a protein, suggesting a mechanism by which the NrdA-a protein might dimerize. Moreover, the observation that we can purify an NrdA-a/NrdA-b complex in the absence of NrdB shows that heterodimerization does not require interactions with NrdB. In considering a potential mechanism by which the two proteins assemble, we noted that the amino acid composition of the NrdA-a and NrdA-b specific ‘tails’ are enriched in charged residues (Fig. 1C). An attractive hypothesis is that the tails facilitate association of NrdA-a and NrdA-b by mediating protein-protein interactions. Interestingly, our data indicate that the split NrdA-a/NrdA-b proteins form a complex with NrdB (Fig. 4B) that has a dissociation constant almost 10-fold stronger than the corresponding E. coli NrdA/NrdB complex (42).
One critical finding was that the split Aeh1 RNR proteins have an activity (≈100 units/mg) similar to that reported for class I RNR α2β2 enzymes that are composed of a contiguous NrdA protein, including the enzyme from the related T-even phage, T4 (SI Table 2). In phage T4, the nrdA gene is not essential, but nrdA mutants have a reduced burst size and a delayed DNA synthesis phenotype compared with wild-type phage (43, 44). The phenotype of nrdA mutant phage suggests a strong selective advantage for T-even phage to possess a functional class I RNR, as wild-type phage would quickly out-compete mutant phage. It is tempting to speculate that the mobE insertion into the Aeh1 nrdA gene may have been tolerated initially because the gene is not essential. We suggest, however, that natural selection would quickly favor phage variants in which a mechanism arose to assemble a functional large subunit from two split halves, because such phage would be at a replicative advantage over nrdA mutant phage. Although the molecular mechanism(s) that promotes assembly of the split RNR proteins remains to be elucidated, it is clear that the mobE insertion is phenotypically neutral with respect to nrdA function, thus minimizing the impact of the insertion on phage viability.
Materials and Methods
Bacteria and Phage.
E. coli DH5α was used for cloning, and E. coli BL21(DE3) was used for protein expression. A. hydrophila C-1 was used for propagation of phage Aeh1 (obtained from the Félix d'Hérelle Reference Center for Bacterial Viruses at Laval University, Quebec, Canada).
Phage Infections.
Overnight cultures of A. hydrophila C-1 were diluted 1/100 in fresh TSB media (EMD BioScience, San Diego, CA) and grown at 30°C to an A600 of 0.4 (≈2 × 108 cells per ml). Aeh1 was infected at a multiplicity of infection of 4–8. For RNA isolation, 3-ml aliquots were removed at various time points. RNA was isolated according to manufacturer's guidelines (Qiagen, Valencia, CA) and treated with TURBO DNase (Ambion, Austin, TX). For protein isolation, 300 ml of A. hydrophila was infected as above, and cells were collected after 20 min and frozen at −80°C.
RT-PCR.
RT-PCR was performed by using 5 μg of total RNA, 20 pmol of primer DE-26, and M-MuLV reverse transcriptase (NEB, Pickering, Canada) in the supplied buffer. A 5-μl aliquot of each reverse transcription reaction served as template for the amplification of the cDNA using Taq DNA polymerase (NEB) in supplied buffer with primers DE-25/DE-26 or DE-143/DE-26.
Protein Purification.
Details of cloning the Aeh1 nrd genes and purification of RNR proteins are available in SI Materials and Methods.
RNR Assays.
In standard assays, proteins were incubated in 50 mM Tris·HCl (pH 7.5)/5 mM ATP/30 mM magnesium acetate/30 mM DTT/0.7 mM [3H]CDP (≈20,000 cpm/pmol) for 20 min at 25°C in a final volume of 50 μl. The amount of dCDP formed was determined by the standard method as described (45). One unit of enzyme corresponds to 1 nmol of dCDP formed per min, and specific activity is units/mg of protein. KD, Vmax, and KL values were obtained by direct curve fitting as described (15, 42) by using KaleidaGraph software (Synergy Software). Before assaying, the NrdB protein was reconstituted at 25°C by reaction with anaerobic ferrous iron ascorbate solution under aerobic conditions as described (46) with a molar ratio of three irons per NrdB polypeptide. After 5 min, the reaction mixture was desalted on a NAP-10 column (GE Healthcare, Piscataway, NJ) equilibrated with buffer B and concentrated with Centricon 30 (Millipore, Bedford, MA).
Mass Spectrometry.
Mass spectrometry was performed at the Functional Proteomics Facility and the Biological Mass Spectrometry Laboratory at the University of Western Ontario. The Aeh1-encoded NrdA-a, NrdA-b, and NrdB proteins were identified by MALDI analyses of proteolytic digestions of excised spots. The A. hydrophlia-encoded NrdA, AAA+ ATPase, and Aeh1-encoded NrdD proteins were identified by Q-TOF MS/MS analyses after separation of peptides by liquid chromatography.
Oligonucleotides.
Sequences of oligonucleotides used in cloning and RT-PCR experiments are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jim Karam (Tulane University, New Orleans, LA) for Aeh1 genomic DNA; Megan Davey and Brent Stead for help with protein purification; MariAnn Westman for help with the enzyme assays; and David Haniford, Niles Lehman, and Marlene Belfort for discussion and reading the manuscript. This work was supported by Natural Sciences and Engineering Research Council of Canada Grant 311610-2005 and Canadian Institutes of Health Research Grant MOP77779 (to D.R.E.) and grants from the Swedish Research Council and Swedish Cancer Foundation to (to B.-M.S.).
Abbreviation
- RNR
ribonucleotide reductase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609915104/DC1.
References
- 1.Belfort M, Derbyshire V, Cousineau B, Lambowitz A. In: Mobile DNA II. Craig N, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: Am Soc Microbiol; 2002. pp. 761–783. [Google Scholar]
- 2.Shub DA, Goodrich-Blair H. Cell. 1992;71:183–186. doi: 10.1016/0092-8674(92)90345-d. [DOI] [PubMed] [Google Scholar]
- 3.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. Annu Rev Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- 4.Lambowitz AM, Zimmerly S. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 5.Sharma M, Ellis RL, Hinton DM. Proc Natl Acad Sci USA. 1992;89:6658–6662. doi: 10.1073/pnas.89.14.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD. Virol J. 2006;3:30. doi: 10.1186/1743-422X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgell DR. In: Homing Endonucleases and Inteins. Belfort M, Stoddard BL, Wood DW, Derbyshire V, editors. Berlin: Springer; 2005. pp. 147–160. [Google Scholar]
- 9.Kutter E, Gachechiladze K, Poglazov A, Marusich E, Shneider M, Aronsson P, Napuli A, Porter D, Mesyanzhinov V. Virus Genes. 1995;11:285–297. doi: 10.1007/BF01728666. [DOI] [PubMed] [Google Scholar]
- 10.Belle A, Landthaler M, Shub DA. Genes Dev. 2002;16:351–362. doi: 10.1101/gad.960302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadyrov FA, Shlyapnikov MG, Kryukov VM. FEBS Lett. 1997;415:75–80. doi: 10.1016/s0014-5793(97)01098-3. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, Belle A, Shub DA, Belfort M, Edgell DR. J Mol Biol. 2003;334:13–23. doi: 10.1016/j.jmb.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 13.Edgell DR, Belfort M, Shub DA. J Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordlund P, Reichard P. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 15.Larsson Birgander P, Bug S, Kasrayan A, Dahlroth SL, Westman M, Gordon E, Sjöberg B-M. J Biol Chem. 2005;280:14997–15003. doi: 10.1074/jbc.M500565200. [DOI] [PubMed] [Google Scholar]
- 16.Landthaler M, Begley U, Lau NC, Shub DA. Nucleic Acids Res. 2002;30:1935–1943. doi: 10.1093/nar/30.9.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu XQ, Yang J, Meng Q. J Biol Chem. 2003;278:46826–46831. doi: 10.1074/jbc.M309575200. [DOI] [PubMed] [Google Scholar]
- 18.Lazarevic V. Nucleic Acids Res. 2001;29:3212–3218. doi: 10.1093/nar/29.15.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy SR, Gold L. Genes Dev. 1991;5:1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen-Lane J, Belfort M. Science. 1987;237:182–184. doi: 10.1126/science.3037701. [DOI] [PubMed] [Google Scholar]
- 21.Sandegren L, Sjöberg B-M. J Biol Chem. 2004;279:22218–22227. doi: 10.1074/jbc.M400929200. [DOI] [PubMed] [Google Scholar]
- 22.Sandegren L, Nord D, Sjöberg B-M. Nucleic Acids Res. 2005;33:6203–6213. doi: 10.1093/nar/gki932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD. J Mol Biol. 2006;361:46–68. doi: 10.1016/j.jmb.2006.05.071. [DOI] [PubMed] [Google Scholar]
- 24.Eriksson M, Uhlin U, Ramaswamy S, Ekberg M, Regnström K, Sjöberg B-M, Eklund H. Structure (London) 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 25.Ekberg M, Sahlin M, Eriksson M, Sjöberg B-M. J Biol Chem. 1996;271:20655–20659. doi: 10.1074/jbc.271.34.20655. [DOI] [PubMed] [Google Scholar]
- 26.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 27.Tan KS, Ong G, Song KP. J Bacteriol. 2005;187:567–575. doi: 10.1128/JB.187.2.567-575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everett KD, Kahane S, Bush RM, Friedman MG. J Bacteriol. 1999;181:4734–4740. doi: 10.1128/jb.181.16.4734-4740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braun V, Mehlig M, Moos M, Rupnik M, Kalt B, Mahony DE, von Eichel-Streiber C. Mol Microbiol. 2000;36:1447–1459. doi: 10.1046/j.1365-2958.2000.01965.x. [DOI] [PubMed] [Google Scholar]
- 30.Berglund O, Eckstein F. Methods Enzymol. 1974;34:253–261. doi: 10.1016/s0076-6879(74)34021-9. [DOI] [PubMed] [Google Scholar]
- 31.Cook KS, Greenberg GR. J Biol Chem. 1983;258:6064–6072. [PubMed] [Google Scholar]
- 32.Torrents E, Eliasson R, Wolpher H, Gräslund A, Reichard P. J Biol Chem. 2001;276:33488–33494. doi: 10.1074/jbc.M103743200. [DOI] [PubMed] [Google Scholar]
- 33.Weiss RB, Huang WM, Dunn DM. Cell. 1990;62:117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang WM, Ao SZ, Casjens S, Orlandi R, Zeikus R, Weiss R, Winge D, Fang M. Science. 1988;239:1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- 35.Parker MM, Belisle M, Belfort M. Genetics. 1999;153:1513–1523. doi: 10.1093/genetics/153.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibb EA, Hausner G. Mycol Res. 2005;109:1112–11126. doi: 10.1017/s095375620500376x. [DOI] [PubMed] [Google Scholar]
- 37.Paquin B, Laforest MJ, Lang BF. Proc Natl Acad Sci USA. 1994;91:11807–11810. doi: 10.1073/pnas.91.25.11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saguez C, Lecellier G, Koll F. Nucleic Acids Res. 2000;28:1299–1306. doi: 10.1093/nar/28.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrov VM, Karam JD. Biochemistry (Moscow) 2004;69:1213–1218. doi: 10.1007/s10541-005-0066-7. [DOI] [PubMed] [Google Scholar]
- 40.Torrents E, Westman M, Sahlin M, Sjöberg B-M. J Biol Chem. 2006;281:25287–25296. doi: 10.1074/jbc.M601794200. [DOI] [PubMed] [Google Scholar]
- 41.Torrents E, Poplawski A, Sjöberg B-M. J Biol Chem. 2005;280:16571–16578. doi: 10.1074/jbc.M501322200. [DOI] [PubMed] [Google Scholar]
- 42.Climent I, Sjöberg B-M, Huang CY. Biochemistry. 1992;31:4801–4807. doi: 10.1021/bi00135a009. [DOI] [PubMed] [Google Scholar]
- 43.Chiu CS, Cox SM, Greenberg GR. J Biol Chem. 1980;255:2747–2751. [PubMed] [Google Scholar]
- 44.Yeh YC, Tessman I. Virology. 1972;47:767–772. doi: 10.1016/0042-6822(72)90567-3. [DOI] [PubMed] [Google Scholar]
- 45.Thelander L, Sjöberg B-M, Eriksson S. Methods Enzymol. 1978;51:227–237. doi: 10.1016/s0076-6879(78)51032-x. [DOI] [PubMed] [Google Scholar]
- 46.Atkin CL, Thelander L, Reichard P, Lang G. J Biol Chem. 1973;248:7464–7472. [PubMed] [Google Scholar]
- 47.Magnani E, Sjölander K, Hake S. Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.