Abstract
The human tripartite motif (TRIM) family comprises 70 members, including HIV restriction factor TRIM5α and disease-associated proteins TRIM20 (pyrin) and TRIM21. TRIM proteins have conserved domain architecture but diverse cellular roles. Here, we describe how the C-terminal PRYSPRY domain mediates diverse TRIM functions. The crystal structure of TRIM21 PRYSPRY in complex with its target IgG Fc reveals a canonical binding interface comprised of two discrete pockets formed by antibody-like variable loops. Alanine scanning of this interface has identified the hot-spot residues that control TRIM21 binding to Fc; the same hot-spots control HIV/murine leukemia virus restriction by TRIM5α and mediate severe familial Mediterranean fever in TRIM20/pyrin. Characterization of the IgG binding site for TRIM21 PRYSPRY reveals TRIM21 as a superantigen analogous to bacterial protein A and suggests that an antibody bipolar bridging mechanism may contribute to the pathogenic accumulation of anti-TRIM21 autoantibody immune complex in autoimmune disease.
Keywords: autoimmunity, familial Mediterranean fever, HIV, TRIM21, TRIM5α
The tripartite motif (TRIM) family of proteins comprises 70 members in the human genome (1), including TRIM21 (Ro52). TRIM proteins are involved in diverse cellular processes, including cell proliferation, differentiation, development, oncogenesis, and apoptosis (1). Within the TRIM family, an expanding group of proteins, TRIM1, TRIM5α, TRIM19, TRIM22, and TRIM32, have also been shown to target retroviruses and prevent their replication inside cells (1). The mechanism of viral restriction is not known, although different TRIMs appear to target different stages of the viral replicative cycle. TRIM5α mediates simian immunodeficiency virus restriction in primates by binding to viral capsid subsequent to cell entry but before reverse transcription (2). Human TRIM5α similarly restricts N-tropic murine leukemia virus (3), but is defective against HIV. Targeting of viral capsid in both TRIM5α and TRIM1 is accomplished through a C-terminal PRYSPRY domain (1, 4). A single mutation in the PRYSPRY domain of human TRIM5α, R332P, is sufficient to confer HIV restriction (5); however, neither the capsid nor its related PRYSPRY binding sites have been defined.
TRIM proteins are multidomain, so-called because of their N-terminal RBCC domains: a RING finger encoding E3 ubiquitin ligase activity (48), a B-box, and a coiled-coil domain mediating oligomerization; however, it is the C-terminal PRYSPRY domain that commonly determines function by acting as a targeting module (Fig. 1 A and B). Mutations in the PRYSPRY domains of several TRIM proteins are associated with disease susceptibility. TRIM20/pyrin mutations give rise to familial Mediterranean fever (FMF), whereas mutations in TRIM18/MID1 cause Opitz G/BBB syndrome (1). Homologous PRYSPRY domains are found in 11 families in the human genome, including the butyrophilins (6). Despite its widespread distribution, very little is known about the PRYSPRY domain or how it has evolved to mediate diverse functions in these proteins.
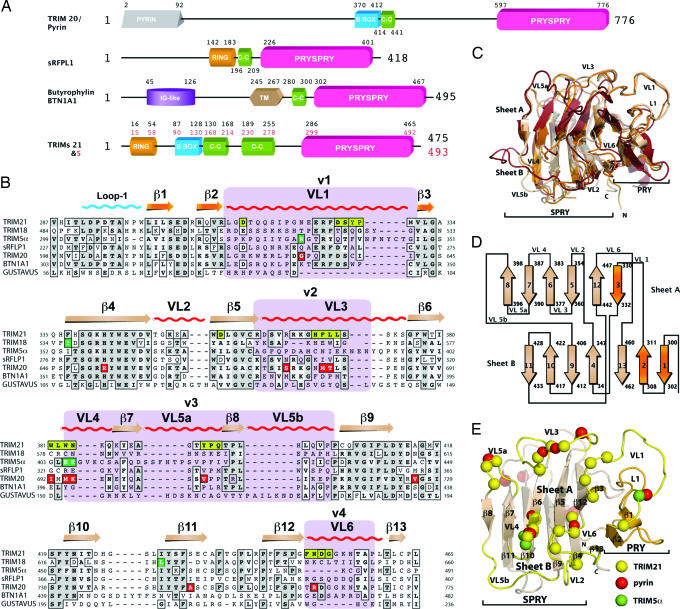
Fig. 1.
The PRYSPRY domain has a conserved binding interface that dictates function in TRIM and other protein families. (A) Domain architecture of TRIM proteins 20, 21, and 5α, Butyrophylin BTN1A1, and putative protein sRFPL1. Domain boundaries as predicted by SMART are indicated (47). (B) Sequence alignment of TRIM21 PRYSPRY with those of disease-related TRIM proteins and homologous SPRY domains. TRIM21 secondary structure elements are indicated along with the position of the canonical binding loops (VLs). TRIM21:Fc contact residues are shown in yellow boxes. Dark green boxes indicate the position of mutations in TRIM5α. Red boxes indicate pyrin mutations that confer FMF susceptibility. The four variable regions known to dictate viral restriction specificity in TRIM5α are marked in purple. Gray shading indicates sequence similarity. (C) Superposition of TRIM21 (wheat), GUSTAVUS (red), and sRFPL1 (orange). (D) PRYSPRY topology cartoon. The PRY subdomain is colored in orange and the SPRY is colored in wheat. (E) TRIM21 PRYSPRY binding site. Variable binding loops are indicated in yellow. Yellow spheres correspond to important contacts with Fc, green indicates TRIM5α mutations, and red shows pyrin mutations linked to FMF.
We chose TRIM21 as a model for understanding PRYSPRY-mediated TRIM function. TRIM21 is a major autoantigen in autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus (SLE), and Sjorgen's syndrome (7–9). Anti-TRIM21 autoantibodies are diagnostic both of disease (10) and disease progression (11), and a pathologic role for TRIM21:autoantibody immune complex has been shown (12). TRIM21 is predominately cytoplasmic (13) but may relocate to the surface of cells during apoptosis (14, 15). Intriguingly, TRIM21 has been shown to bind both normal serum IgG and anti-TRIM21 autoantibodies (16, 17). Immunoprecipitation studies suggest that IgG binding is mediated by the PRYSPRY domain (18), but how it targets IgG and how this might contribute to disease is not known.
We have determined the crystal structure of TRIM21 PRYSPRY bound to IgG Fc, revealing how TRIM PRYSPRY domains bind their target. The structure defines the canonical interface for the PRYSPRY superfamily and shows that, contrary to prediction, it is comprised of two discrete binding pockets. Alanine scanning reveals that conserved hot-spot positions within the interface dictate the function of TRIM20/pyrin, TRIM5α, and TRIM21. Finally, characterization of the IgG binding site of TRIM21 PRYSPRY suggests that anti-TRIM21 autoantibodies may undergo antibody bipolar bridging, preventing efficient immune complex clearance and contributing to pathogenic accumulation in disease.
Results
TRIM PRYSPRY Domains Have a Canonical Binding Interface.
The TRIM21 PRYSPRY:Fc complex was solved by molecular replacement in P61 to 2.35-Å resolution [supporting information (SI) Table 1]. TRIM21 PRYSPRY forms a single globular fold comprising a distorted β-sandwich of two antiparallel β-sheets similar to the “PRYSPRY” structure of sRFPL1, a protein of unknown function (19) and the SPRY domains of SOCS box proteins SSB and GUSTAVUS (Fig. 1 C–E). There are two TRIM21 PRYSPRY molecules per asymmetric unit, each of which binds to one chain of the Fc homodimer (Fig. 2A). The binding interface is formed by six extended loops that are analogous to the six CDR loops in antibodies and that we have labeled variable loops (VLs) (Fig. 1). Sequence alignment of homologous PRYSPRY domains shows that these binding site loops define regions of hypervariability (Fig. 1B). Conserved residues map to the hydrophobic core between the two β-sheets and to secondary structure elements, suggesting that TRIM PRYPSRY domains have a canonical binding interface and that new functions evolve by loop diversification. The location of the binding interface in TRIM21 is similar to the dimerization interface observed in sRFPL1 (19). Based on their uncomplexed structure of GUSTAVUS, Woo et al. (20) predicted two SPRY interaction surfaces at opposite ends of the domain; however, neither surface matches the Fc binding site on TRIM21. The TRIM21 complex reveals that the six VLs create two distinct binding pockets, in contrast to predictions based on previous structures (Fig. 2 B and C) (19–21). These two binding pockets locate to the separate “PRY” and “SPRY” subdomains, suggesting that although they form a single interface and structural fold they may encode discrete functional units.
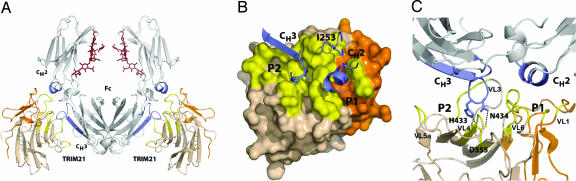
Fig. 2.
The TRIM21 PRYSPRY:Fc complex. (A) Secondary structure representation of the complex. Contact areas on the Fc are marked in dark blue and on TRIM21 in yellow. The PRY subdomain of TRIM21 is marked in orange and SPRY in wheat. (B) Surface representation of the complex showing the two binding pockets (P1 and P2) and contacting Fc structure (blue). Yellow indicates surface buried upon complex formation. (C) Secondary structure representation of the TRIM21:Fc interface showing two discrete pockets and hydrogen-bond interactions between Fc residues H433 and N434 and TRIM21 D355.
In TRIM21, the PRY subdomains form a binding pocket for the first Ig domain of Fc, CH2, whereas the SPRY subdomains form a second binding pocket for the second Ig domain, CH3 (Fig. 2 B and C). The PRY-encoded binding pocket is entirely novel and was not previously predicted, possibly because VL1 and VL6, from which it is comprised, constitute the most variable regions between PRYSPRY homologues. In TRIM21, residues from VL1 and VL6 create a pocket for CH2 α-helix310–314 and for I253 from loop243–258 (Fig. 2 B and C). I253 is one of six hot-spot residues identified by de Lano et al. (22) as critical for proteins that bind to the CH2-CH3 Fc interface. In addition to I253, TRIM21 binds to three other Fc hot-spot residues, namely H433, N434, and H435.
The SPRY subdomain forms a large enclosed pocket in which residues from VLs 2–6 and β-strands 5 and 6 interact with Fc CH3 residues from the two N-terminal β-strands and connecting loop428–436 (Fig. 2 B and C). Fc loop428–436 penetrates deep into the binding pocket where apex residues H433Fc and N434Fc form a hydrogen-bond network with D355TR (Fig. 2C). Together, the PRY and SPRY binding pockets provide a complementary interface for IgG Fc with a shape complementarity score (Sc score) of 0.68, similar to protein A:Fc, which has a score of 0.66 [Sc scores were calculated by using the program Sc in CCP4 (23)]. This Sc number is normal for an IgG interface but not high when compared with dimeric interfaces (values are ≈0.75). A ring of nonpolar residues borders the interface to create a solvent-excluded hydrophobic surface with a buried surface area of 1,557 Å2.
TRIM21 and Fc Hot-Spot Residues Colocate.
To determine which residues in the TRIM21 PRYSPRY interface are critical for binding, we mutated those whose side chains contact Fc. Each mutant was expressed and purified as with WT, its association and dissociation kinetics were determined by stopped-flow, and its steady-state affinity was measured by fluorescence anisotropy (SI Table 2). Fig. 3 shows that side-chain interactions from residues distributed across the interface contribute to binding. Of the 13 mutants tested, H368A and Y328F increased binding by a ΔΔG of −0.9 and −1.6 kcal/mol, respectively. Mutant Y328A shows decreased binding (ΔΔG = 1 kcal/mol), suggesting that a side chain at this position is required for hydrophobic stacking but not hydrogen bonding. Why H368A increases binding is less clear, although it is structurally adjacent to Y328 and mutation to alanine may prevent the histidine competing with the position occupied by Y328 to provide a less suitable stacking partner.
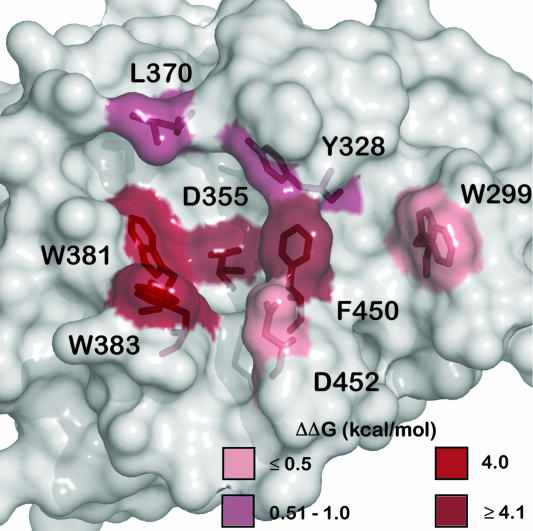
Fig. 3.
Alanine scan of the TRIM21 PRYSPRY binding interface. Mutants that reduce affinity are indicated in red, the darker the color the greater the effect on the ΔΔG of binding.
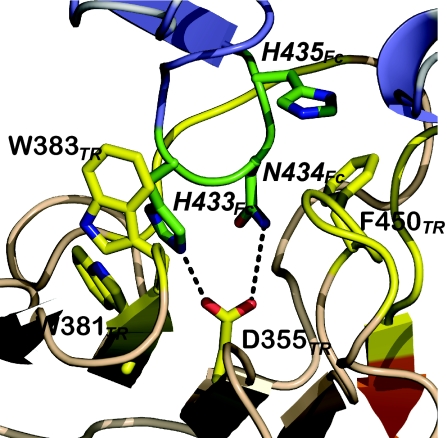
Four TRIM21 hot-spot residues (D355, W381, W383, and F450) are absolutely required for Fc binding, and mutation of any single residue effectively abolishes interaction (ΔΔG ≥4 kcal/mol). These four residues are clustered at the center of the interface and contact the three Fc hot-spot residues in loop433–436 (H433Fc, N434Fc, and H435Fc) (Fig. 4). This colocalization of hot spots substantiates our proposal that interaction between Fc and TRIM21 is specific and conserved. Taken together with the crystal structure, these data reveal a hydrophobic pocket at the center of the interface in which both hydrophobic stacking interactions and a central hydrogen-bond network provide the energy for binding. A list of all TRIM21:Fc interactions is given in SI Data Set.
Fig. 4.
TRIM21 PRYSPRY hot-spot residues bind Fc hot-spot residues. Secondary structure representation of the core binding interactions; TRIM21 (TR) is in orange with yellow side chains; Fc is in blue with green side chains.
TRIM21 PRYSPRY Associates with IgG in a Single Step with Fast Association Kinetics and High Affinity.
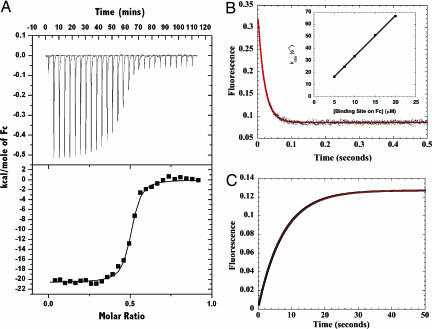
Isothermal titration calorimetry (ITC) experiments show that binding of TRIM21 to IgG Fc occurs with high affinity (37 nM) and confirms a stoichiometry of two molecules of TRIM21 to one Fc fragment (Fig. 5A). Measurement under different buffer conditions reveals that binding is salt-dependent but pH-independent. Varying the pH from pH 8 to pH 5 does not significantly affect binding; however, a 10-fold reduction in salt concentration from 200 to 20 mM results in a 5-fold increase in affinity, manifested in a decreased off-rate (data not shown). Binding of Fc by TRIM21 is therefore similar to superantigens protein A and G, in contrast to the pH-dependent binding of the neonatal Fc receptor FcRn and herpes simplex virus Fc receptor gE-gI (24, 25). Binding is enthalpically driven (ΔH =−20.6 kcal·mol−1) and entropically unfavorable (ΔS =−102 cal·mol−1·K−1) which, together with a strongly negative (−900 cal·mol−1·K−1) ΔCp (dΔH/dT), can indicate that binding involves concomitant structural change (26).
Fig. 5.
Kinetic and thermodynamic analysis of the TRIM21 PRYSPRY domain binding to the IgG Fc region. (A) Titration of TRIM21 into IgG Fc measured by ITC shows that two molecules of TRIM21 bind to each IgG Fc with a Kd of 37 nM. (B) Rapid mixing of TRIM21 with IgG Fc in a stopped-flow experiment results in a fast monophasic fluorescence quench. The data are shown as black dots with the fit to a single exponential function shown as a solid red line. (Inset) A linear increase in kobs with increasing pseudofirst-order concentration of Fc indicates a single step binding mechanism. Linear regression yields kon = 3.5 × 106 M−1·s−1. (C) Mixing of excess protein A outcompetes TRIM21 for binding to IgG Fc resulting in a fluorescence enhancement with an observed rate kdiss of 0.13 s−1 to yield a kinetic Kd (= kdiss/kon) of 37 nM.
To determine whether the TRIM21 PRYSPRY binding site is preformed or undergoes structural rearrangement we followed the presteady-state binding kinetics by stopped-flow, taking advantage of the quench in tryptophan fluorescence accompanying binding (Fig. 5B). We measured a rapid monophasic fluorescence quench with a fast bimolecular association rate constant (kon) of 3.5 × 106 M−1·s−1 and a dissociation rate constant slower than 0.2 s−1 (the small value precluding more accurate measurement). This single bimolecular step suggests that both TRIM21 and Fc binding sites are preformed and that binding to one Fc site does not influence binding to the second.
The bacterial B cell superantigen proteins A and G produced by Staphylococcus aureus and Streptococcal species, respectively, have previously been shown to bind with high (nanomolar) affinity to the CH2–CH3 interface of Fc (27, 28). We used the fluorescence-neutral binding of protein A to determine an accurate TRIM21:Fc dissociation rate constant in a chase experiment. Mixing preformed TRIM21:Fc complex (2.5 μM TRIM21 and 1 μM Fc) with excess (25 μM) protein A in a stopped-flow gave a slow fluorescence enhancement corresponding to a koff of 0.13 s−1 (Fig. 5C). Similar results were obtained with protein G but with a much smaller amplitude change (data not shown). This experiment confirms the structural prediction that TRIM21 competes for the same CH2–CH3 site as protein A/G. The bimolecular microscopic rate constants predict a steady-state affinity of 37 nM, similar to the Kd determined by ITC experiments, confirming that TRIM21:Fc binding occurs in a single collision step. Together with the alanine scan data showing coalignment of hot spot to hot spot, these kinetic data support our hypothesis that Fc binding is both evolved and specific. The single binding step allows rapid association with Fc; fast association kinetics may be important in antiviral TRIM proteins such as TRIM5α, which must associate with HIV capsid immediately before uncoating.
Discussion
The PRYPSRY Domain Is a Versatile Modular Scaffold that Readily Incorporates New Functions.
The PRYSPRY domain is a common protein module found in 11 families in the human genome (6), including the functionally diverse TRIM family. Three structures of SPRY-containing molecules have been presented recently, revealing a compact β-fold similar to the galectins (19–21). The TRIM21 PRYSPRY:Fc structure described here is a complexed structure involving the PRYSRY domain showing that it forms a canonical binding interface. Within this interface, the PRY and SPRY subdomains each encode a discrete binding pocket. We have classified six antibody-like VLs that determine this interface. Two of these VLs, VL1 and VL6, lie outside what is commonly considered the core domain (21), yet they play a critical role in TRIM21:Fc binding and greatly enhance the possibility of combinatorial diversity of the PRYSPRY binding site. The antibody-like binding site architecture of the PRYSPRY domain suggests that it is capable of binding targets ranging from small molecules and linear epitopes such as peptides to discontinuous conformational epitopes provided by proteins (as illustrated by the TRIM21:Fc structure).
Despite significant sequence variation, there are structural similarities between TRIM21 and the PRYSPRY domain of predicted protein sRFPL1 (Fig. 1C). The main-chain conformations of binding site loops VL1–VL3 and VL5 closely align (Cα rmsd ≈1 Å,) with only the hot-spot loops VL4 and VL6 showing significant variation (Cα rmsd of 2 and 5 Å, respectively). Notwithstanding these variations, sRFPL1 recapitulates much of the topography of the Fc binding site of TRIM21 (SI Fig. 6). To test whether sRFPL1 binds Fc we expressed both the crystallized construct and a construct in which the C terminus that mediates dimerization is deleted. Neither construct bound to IgG or Fc as measured by ITC, gel filtration, or anisotropic fluorescence. This finding suggests that although the binding sites of sRFPL1 and TRIM21 are similar there are functionally significant differences.
It is possible that TRIM proteins adopt a small number of canonical loop conformations analogous to antibodies. TRIM VL loop sequences are highly divergent, but they are constrained by the surrounding scaffold, which is well conserved. The VL1 loops of sRFPL1 and TRIM21 adopt very similar backbone conformations because of constraining interactions mediated by conserved loop-1 residues LDPXTA and VL1 residues RF(D). Alignment of all TRIM PRYSPRY domains (data not shown) reveals that >90% contain elements of this loop-1 consensus sequence, whereas most contain the VL1 consensus motif. Thus, we predict that the conformation of VL1 may be conserved in other TRIM PRYSPRY domains. In TRIM21, loop-1 consensus residues initiate a tight β-turn to create a loop apex around which VL1 lassoes. VL1 is held in place by consensus residue R324, which hydrogen-bonds with the main-chain oxygen of loop-1 A296 through its Nε atom and the peptide oxygen of P293 through its amino group. VL1 is further constrained against VL3 and β-strand 3 by consensus residues F325 and D326, respectively. Sequence alignment of the human PRYSPRY families reveals that the aspartic acid of the LDPXTA motif is preserved in 8 of 11 families (data not shown). PRYSPRY families from other species, such as Enterophilin, Stonustoxin, and Ohanin, also contain the core LDP and RF(D) motifs (data not shown). Conservation of these motifs suggests that a significant degree of structural homology can be expected in this region of the molecule.
TRIM21 PRYSPRY Is a Human B Cell Superantigen.
Comparison of TRIM21 to other Fc binding proteins shows that it binds at a different location to mammalian Fcγ receptors. Most mammalian Fcγ receptors bind on or near the top of the CH2 domain; only the neonatal receptor FcRn does not (29). FcRn binds both to the CH2–CH3 interface and the CH3 domain via an N-linked carbohydrate attached to N128 (29). Rheumatoid factors (anti-IgG antibodies commonly detected during rheumatoid arthritis) also bind the CH2–CH3 domain interface. However, the structure of a rheumatoid factor bound to Fc reveals that binding occurs with poor complementarity and uses only the very edge of the antibody combining site (30). This adventitious interaction contrasts with TRIM21:Fc, in which all of the features of the binding interface suggest an evolved interaction. Binding to the CH2–CH3 interface is a feature of pathogen-derived B cell superantigens, such as protein A (31), protein G (32), and herpes simplex virus 1 gE-gI (33). There are several physiological implications to binding in this way. First, as the CH2–CH3 interface is highly conserved, it facilitates binding by TRIM21 to >98% of circulating antibodies regardless of their antigen specificity. Second, the TRIM21 Fc-binding site likely does not overlap with the binding sites for FcγR and C1q. Thus binding of TRIM21 to nonautoantibody IgG probably does not inhibit Fcγ-mediated mechanisms such as antibody-dependent cellular cytoxicity and complement recruitment and is unlikely to be pathogenic.
Autoantibody Bipolar Bridging.
TRIM21 is involved in several autoimmune diseases such as SLE. Although the pathogenicity of TRIM21:nonanti-TRIM antibody complexes has not been directly tested, we believe that such binding is unlikely to be pathogenic as TRIM21 is continually exposed to the immune system either after necrotic cell death or by locating to the outside of cells during apoptosis (14, 34). Furthermore, TRIM21:serum IgG complex is likely to be cleared via complement and Fcγ receptors. However, during autoimmune diseases such as SLE, anti-TRIM21 autoantibodies are generated whose Fabs bind epitopes in the RING and B-box domains of TRIM21 (35). Based on our data, we predict that these autoantibodies will also bind to the PRYSPRY domain via their Fc. The simultaneous binding of both Fab and Fc, or “antibody bipolar bridging,” is a feature of pathogen superantigens protein A, protein G, and gE-gI (33, 36). Proteins A and G possess multiple domains that permit binding both to the same region on Fc as TRIM21 and an additional epitope on the Fab. Antibody bipolar bridging is thought to occlude the binding site on CH2 for Fcγ receptor and complement, disconnecting antibody recognition and effector function and facilitating immune evasion. We propose that anti-TRIM21 autoantibodies undergo TRIM21-mediated bipolar bridging, resulting in their cross-linking and a similar block to Fcγ receptor and complement interaction. TRIM21-mediated bipolar bridging could occur either in cis, a single IgG bound by a single TRIM molecule, or, more likely, in trans, the N terminus of one TRIM molecule interacting with an anti-TRIM21 autoantibody that is bound by the PRYSPRY domain of a second TRIM molecule. TRIM21 is trimeric, and each autoantibody has four potential binding sites, (two on the Fab and two on the Fc), therefore it is likely that the resulting cross-linked bipolar-bridged immune complexes form very large protein aggregates.
Autoantibody bipolar bridging could affect autoimmune pathogenesis in several ways. First, bipolar-bridged complexes may contribute to the pathogenic deposition of immune complex in SLE (37). If formation of TRIM21 cross-linked autoantibody aggregates blocked the binding of Fcγ receptor and complement, immune complexes would not be cleared from the serum, in contrast to TRIM21:nonautoantibody complexes. Second, bipolar-bridged autoantibodies associated with TRIM21 on the surface of apoptosing cells could inhibit cell clearance, for example by blocking access to so-called “eat me” signals on the cell surface (38). This hypothesis could explain the defective clearance of apoptosed cells observed in diseased SLE mice (39).
A Structural Explanation for TRIM5α and TRIM20/Pyrin Mutations.
Characterization of the PRYSPRY binding interface helps to explain many of the mutations known to affect function in disease-associated TRIM proteins. Mutations in TRIM5α PRYSPRY dictate viral restriction specificity and efficacy (40), and on the basis of sequence variation between primates, four variable regions v1–v4 have been identified (4). These regions map closely to our VL definitions (Fig. 1 B and D), suggesting that they directly mediate interaction with viral epitopes. Human TRIM5α is defective in HIV restriction but effective against murine leukemia virus (MLV). Restriction is specific to the N-tropic form of MLV and depends on PRYSPRY residues E405 and E406 (41). These residues map directly onto Fc-contacting residues in TRIM21 (Fig. 1 B and E). The ability to restrict HIV can be conferred on human TRIM5α by a single mutation, R332P (5). The TRIM21 structure shows that this residue locates to the unpredicted PRY binding pocket formed by VL1 (Fig. 1 B and E) and suggests that it is involved in capsid recognition.
TRIM20/Pyrin is a disease-associated TRIM protein in which PRYSPRY mutations are correlated with susceptibility to FMF (42). Most of these mutations map onto the binding loops of TRIM21 (Fig. 1 B and E). In particular, pyrin mutations in VL3–6 exactly map to critical Fc-contact residues in TRIM21. Of these, the VL3 and VL4 mutations (M680L, T681I, and M694V/L/I) give rise to the most severe FMF symptoms described (43). Mutations I692del and M694V/L/I map directly onto TRIM21 hot-spot residues W381TR and W383TR, whereas R761H is adjacent to hot-spot residue F450TR. Thus TRIM21, TRIM5α, and TRIM20/pyrin all share hot spots around position 383TR, suggesting that it forms a conserved hot spot within the canonical PRYSPRY interface (Fig. 3). Finally, pyrin has a disease-associated polymorphism (G632S) in VL1 that is structurally equivalent to mutation R332P in TRIM5α, suggesting that it may also be a common hot-spot position. Together, these data suggest that TRIM PRYSPRY function is heavily influenced by a few conserved hot-spot positions.
In summary, PRYSPRY domains mediate diverse functions by high-affinity binding to their target through a canonical binding interface formed by six antibody-like VLs. We propose that diversification of these six VLs has driven the rapid evolution of PRYSPRY function. Within the TRIM family, conserved structural features and hot-spot positions within these loops dictate the binding of autoantigen TRIM21 to IgG Fc and the targeting of retroviral capsid by TRIM5α and determine disease severity in pyrin/FMF.
Materials and Methods
Protein Expression and Purification.
TRIM21 and sRFPL1 PRYSPRY domain was expressed in BL21-derived cells under standard conditions (see SI Text for details) (44).
Rapid Reaction Kinetics.
Experiments were carried out largely as described (see SI Text for details) (44). Data were collected at 30°C in an SX-18MV stopped-flow spectrofluorimeter (Applied Photophysics, Leatherhead, U.K.) using 1 μM TRIM21 and increasing pseudofirst-order concentrations of Fc.
Titration Calorimetry and Fluorescence Anisotropy.
Experiments were carried out largely as described (see SI Text for details) (45). ITC experiments were carried out by using a VP-ITC (Microcal, Amherst, MA) with 10 μM of TRIM21 in the cell and 60 μM Fc in the syringe. Anisotropy experiments were carried out by using a LS-50b luminescence spectrofluorimeter (PerkinElmer, Wellesley, MA) on 1 μM TRIM21 and increasing pseudofirst-order concentrations of IgG.
Crystallography.
Data from complexed crystals were collected at European Synchrotron Radiation Facility beamline ID23-1 (Grenoble, France) and phased by molecular replacement using PHASER (46). For a complete description see SI Text.
Supplementary Material
Acknowledgments
We thank Didier Nurizzo for assistance at European Synchrotron Radiation Facility beamline ID23-2. L.C.J. thanks Dr. Roger Williams and Professor Stanley Fields for critical reading of the manuscript. L.C.J., A.H.K. and Z.K. were supported by the Medical Research Council, and D.A.R. and J.T. were supported by a grant from the Wellcome Trust.
Abbreviations
- TRIM
tripartite motif
- FMF
familial Mediterranean fever
- SLE
systemic lupus erythematosus
- VL
variable loop
- ITC
isothermal titration calorimetry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2IWG).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609174104/DC1.
References
- 1.Nisole S, Stoye JP, Saib A. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 2.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. Proc Natl Acad Sci USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song B, Gold B, O'Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. J Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yap MW, Nisole S, Stoye JP. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes DA, de Bono B, Trowsdale J. Immunology. 2005;116:411–417. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Chetrit E, Chan EK, Sullivan KF, Tan EM. J Exp Med. 1988;167:1560–1571. doi: 10.1084/jem.167.5.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Chetrit E, Fox RI, Tan EM. Arthritis Rheum. 1990;33:349–355. doi: 10.1002/art.1780330307. [DOI] [PubMed] [Google Scholar]
- 9.Moutsopoulos HM, Skopouli FN, Sarras AK, Tsampoulas C, Mavridis AK, Constantopoulos SH, Maddison PJ. Ann Rheum Dis. 1985;44:215–219. doi: 10.1136/ard.44.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCauliffe DP, Wang L, Satoh M, Reeves WH, Small D. J Rheumatol. 1997;24:860–866. [PubMed] [Google Scholar]
- 11.Frank MB, Itoh K, Fujisaku A, Pontarotti P, Mattei MG, Neas BR. Am J Hum Genet. 1993;52:183–191. [PMC free article] [PubMed] [Google Scholar]
- 12.Salomonsson S, Sonesson SE, Ottosson L, Muhallab S, Olsson T, Sunnerhagen M, Kuchroo VK, Thoren P, Herlenius E, Wahren-Herlenius M. J Exp Med. 2005;201:11–17. doi: 10.1084/jem.20041859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes DA, Ihrke G, Reinicke AT, Malcherek G, Towey M, Isenberg DA, Trowsdale J. Immunology. 2002;106:246–256. doi: 10.1046/j.1365-2567.2002.01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casciola-Rosen LA, Anhalt G, Rosen A. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohlsson M, Jonsson R, Brokstad KA. Scand J Immunol. 2002;56:456–469. doi: 10.1046/j.1365-3083.2002.01072_79.x. [DOI] [PubMed] [Google Scholar]
- 16.Yang YS, Yang MC, Wang B, Weissler JC. Mol Immunol. 2000;37:591–602. doi: 10.1016/s0161-5890(00)00068-7. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, Eversole T, Lee DJ, Sontheimer RD, Capra JD. Scand J Immunol. 1999;49:620–628. doi: 10.1046/j.1365-3083.1999.00547.x. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes DA, Trowsdale J. Mol Immunol. 2007;44:2406–2414. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Grutter C, Briand C, Capitani G, Mittl PR, Papin S, Tschopp J, Grutter MG. FEBS Lett. 2006;580:99–106. doi: 10.1016/j.febslet.2005.11.076. [DOI] [PubMed] [Google Scholar]
- 20.Woo JS, Imm JH, Min CK, Kim KJ, Cha SS, Oh BH. EMBO J. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters SL, Yao S, Willson TA, Zhang JG, Palmer KR, Smith BJ, Babon JJ, Nicola NA, Norton RS, Nicholson SE. Nat Struct Mol Biol. 2006;13:77–84. doi: 10.1038/nsmb1034. [DOI] [PubMed] [Google Scholar]
- 22.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 23.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Raghavan M, Bonagura VR, Morrison SL, Bjorkman PJ. Biochemistry. 1995;34:14649–14657. doi: 10.1021/bi00045a005. [DOI] [PubMed] [Google Scholar]
- 25.Sprague ER, Martin WL, Bjorkman PJ. J Biol Chem. 2004;279:14184–14193. doi: 10.1074/jbc.M313281200. [DOI] [PubMed] [Google Scholar]
- 26.Stites WE. Chem Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 27.Walker KN, Bottomley SP, Popplewell AG, Sutton BJ, Gore MG. Biochem J. 1995;310:177–184. doi: 10.1042/bj3100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson R, Jendeberg L, Nilsson B, Nilsson J, Nygren PA. J Immunol Methods. 1995;183:43–49. doi: 10.1016/0022-1759(95)00030-e. [DOI] [PubMed] [Google Scholar]
- 29.Martin WL, West AP, Jr, Gan L, Bjorkman PJ. Mol Cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 30.Corper AL, Sohi MK, Bonagura VR, Steinitz M, Jefferis R, Feinstein A, Beale D, Taussig MJ, Sutton BJ. Nat Struct Biol. 1997;4:374–381. doi: 10.1038/nsb0597-374. [DOI] [PubMed] [Google Scholar]
- 31.Deisenhofer J. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 32.Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA. Structure (London) 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 33.Sprague ER, Wang C, Baker D, Bjorkman PJ. PLoS Biol. 2006;4:e148. doi: 10.1371/journal.pbio.0040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawley W, Doherty A, Denniss S, Chauhan D, Pruijn G, van Venrooij WJ, Lunec J, Herbert K. Rheumatology (Oxford) 2000;39:253–261. doi: 10.1093/rheumatology/39.3.253. [DOI] [PubMed] [Google Scholar]
- 35.Ottosson L, Hennig J, Espinosa A, Brauner S, Wahren-Herlenius M, Sunnerhagen M. Mol Immunol. 2006;43:588–598. doi: 10.1016/j.molimm.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Leung-Tack J, Neveu T, Lefroit-Joliy M, Voisin GA. Immunol Lett. 1982;5:23–28. doi: 10.1016/0165-2478(82)90086-4. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan KE, Jawad AF, Piliero LM, Kim N, Luan X, Goldman D, Petri M. Rheumatology (Oxford) 2003;42:446–452. doi: 10.1093/rheumatology/keg157. [DOI] [PubMed] [Google Scholar]
- 38.Gardai SJ, Bratton DL, Ogden CA, Henson PM. J Leukocyte Biol. 2006;79:896–903. doi: 10.1189/jlb.1005550. [DOI] [PubMed] [Google Scholar]
- 39.Licht R, Dieker JW, Jacobs CW, Tax WJ, Berden JH. J Autoimmun. 2004;22:139–145. doi: 10.1016/j.jaut.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. J Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perron MJ, Stremlau M, Sodroski J. J Virol. 2006;80:5631–5636. doi: 10.1128/JVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.TIF Consortium. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 43.Goulielmos GN, Fragouli E, Aksentijevich I, Sidiropoulos P, Boumpas DT, Eliopoulos E. Biochem Biophys Res Commun. 2006;345:1326–1332. doi: 10.1016/j.bbrc.2006.04.185. [DOI] [PubMed] [Google Scholar]
- 44.James LC, Roversi P, Tawfik DS. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proc Natl Acad Sci USA. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 47.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freemont PS. Curr Biol. 2000;10:84–87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.